GENETEX - A GENomics Report TEXt Mining R Package and Shiny Application Designed to Capture Real-World Clinico-Genomic Data
Metrics
Journal: JAMIA Open. Published September 28, 2021
Article type: Application Note
Authors: David M. Miller1,2^* MD PhD, Sophia Z. Shalhout1 PhD
1Department of Medicine, Division of Hematology/Oncology and the 2Department of Dermatology, Massachusetts General Hospital, Boston, MA
*Corresponding author: David M. Miller MD PhD
Massachusetts General Hospital
15 Parkman St. Room 132
Boston MA 02114
Funding sources: The Harvard Cancer Center Merkel Cell Carcinoma patient registry is supported by grants from Project Data Sphere, ECOG-ACRIN and the American Skin Association.
Conflicts of interest: None
Keywords: clinico-genomics, data abstraction, electronic health records, shiny app, REDCap, clinical informatics
Acknowledgments: We would like to acknowledge Ravikumar Komandur, PhD, Project Director at Project Data Sphere for review and critique of the manuscript; Guardant Health and Foundation Medicine for making sample reports available for the development of this package. GENETEX is for research purposes only. No clinical decisions should be made with the information obtained from its output. This article reflects the views and work (including development and use of GENETEX) of the authors and should not be construed to represent the work, policies, of any of the vendors whose reports were used to develop GENETEX and whose reports may be provided as part of the package.
Abbreviations: BWH: Brigham and Women’s hospital, CGD: clinico-genomic data, CNV: copy number variants, EDC: electronic data capture, EHR: electronic health record, MGH: Massachusetts General Hospital, MMR: mismatch repair, NSLS: numeric suffix linker system, REDCap: Research Electronic Data Capture, RWD: real-world data, TMB: tumor mutation burden
ABSTRACT
Objectives: Clinico-Genomic Data (CGD) acquired through routine clinical practice has the potential to improve our understanding of clinical oncology. However, these data often reside in heterogeneous and semi-structured data, resulting in prolonged time-to-analyses.
Materials and Methods: We created GENETEX: an R package and Shiny application for text mining genomic reports from EHR and direct import into REDCap®.
Results: GENETEX facilitates the abstraction of CGD from EHR and streamlines capture of structured data into REDCap®. Its functions include natural language processing of key genomic information, transformation of semi-structured data into structured data and importation into REDCap. When evaluated with manual abstraction, GENETEX had >99% agreement and captured CGD in approximately one-fifth the time.
Conclusions: GENETEX is freely available under the Massachusetts Institute of Technology license and can be obtained from GitHub(https://github.com/TheMillerLab/genetex). GENETEX is executed in R and deployed as a Shiny application for non-R users. It produces high-fidelity abstraction of CGD in a fraction of the time.
Lay Summary:
Advances in clinical oncology require a deep understanding of cancer biology. Data regarding the underlying genetic alterations in cancers obtained from routine clinical practice can greatly increase our comprehension of tumor biology. However, there are a number of barriers that impede capitalization of these critical real-world data. Paramount amongst these obstacles are prolonged time-to-analyses secondary to the difficulties of capturing data from heterogeneous sources, as well as the challenges of processing vast amounts of genomic information. These hurdles increase time-to-insight from these data and threaten our ability to fully maximize on advances in molecular and information technologies. In the real-world setting, genomic information resides in a variety of formats. The most common is a report from an institutional molecular pathology department or a commercial vendor. Manual abstraction of these data can be resource intensive. In order to facilitate the abstraction of these data, we created a user-friendly application called GENETEX. GENETEX utilizes natural language processing techniques, such as text mining, to efficiently capture and store genomic information acquired during routine clinical practice. Capturing clinical genomic data in an efficient and structured format can improve analysis and potentially lead to novel insights.
Objectives
Advances in clinical oncology require a deep understanding of cancer biology. Clinico-Genomic Data (CGD) obtained from routine clinical practice can greatly increase our comprehension of tumor biology. However, there are a number of barriers that impede capitalization of these critical real-world data (RWD). Paramount amongst these obstacles include prolonged time-to-analyses secondary to the difficulties of capturing data from heterogeneous sources, as well as the challenges of processing vast amounts of genomic information. These hurdles increase time-to-insight from RWD and threaten our ability to fully maximize on advances in molecular and information technologies.
In the real-world setting, genomic information resides in a variety of formats. The most common is a report from an institutional molecular pathology department or a commercial vendor. This information is often accessed by clinicians or clinical researchers as semi-structured data. Collecting CGD of a patient cohort in a structured electronic data capture (EDC) system can facilitate analysis and reduce time-to-analytics and time-to-action. CGD are often captured via classical (i.e. manual) abstraction by individual research teams. Easily adoptable methods of large-scale CGD collection are limited but include direct transfer from individual vendors. These types of data transfer often require collaborative agreements between vendors and end-users (e.g. investigators/institutions), which can limit their scalability.
We previously published an overview of a methodology and design of a Research Electronic Data Capture (REDCap®)1-based system to facilitate capture of RWD2. REDCap® is a web-based EDC utilized by researchers to collect structured data1. That platform incorporates a form entitled Genomics Instrument, which provides a structured format for the collection of CGD3. This instrument is freely available and can be incorporated into any existing REDCap project. It is currently being used by the Project Data Sphere led Merkel Cell Carcinoma Patient Registry4 to capture CGD.
Here we present GENETEX (pronounced “genetics”), an R package with a Shiny application front-end, which facilitates the abstraction of CGD from EHR and streamlines capture of structured data into the Genomics Instrument in REDCap®. Its functions include natural language processing of key genomic information, transformation of semi-structured data into structured data, and importation into REDCap® (Figure 1). GENETEX is executed in R but is deployed as a Shiny application to enhance the user interface for non-R users.
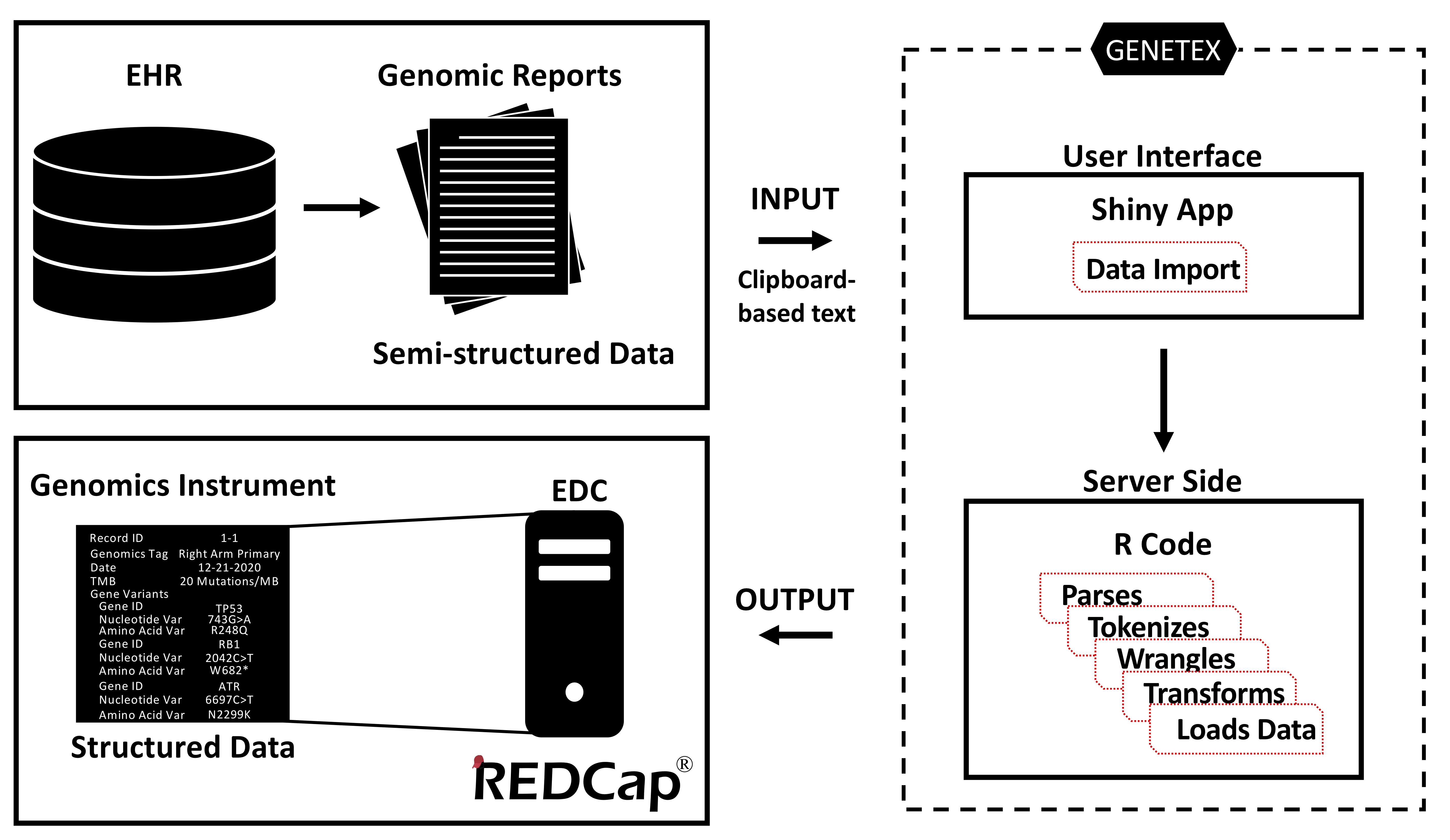 Figure 1. Schema of GENETEX The GENETEX package takes clinico-genomic data, which is typically stored in semi-structured data, as an input via a Shiny application user interface. Once the input data has been captured, the package executes a series of server-side functions that text mine CGD reports for relevant genomic data. These structured data are then imported directly into the REDCap electronic data capture system (EDC), placing the data in the Genomics Instrument in REDCap.
Figure 1. Schema of GENETEX The GENETEX package takes clinico-genomic data, which is typically stored in semi-structured data, as an input via a Shiny application user interface. Once the input data has been captured, the package executes a series of server-side functions that text mine CGD reports for relevant genomic data. These structured data are then imported directly into the REDCap electronic data capture system (EDC), placing the data in the Genomics Instrument in REDCap.
Methods
Software Dependencies
GENETEX is written in R (version 4.0.0), organized using roxygen25, and utilizes the following packages dplyr6, tidyr7, readr8, stringr9, purrr10, REDCapR11, magrittr12, splitstackshape13 and Shiny14. For full details, instructions and examples refer to either our README (https://github.com/TheMillerLab/genetex/blob/main/README.md) or video demonstration (https://github.com/TheMillerLab/genetex/blob/main/Demo_Video.md), both of which can be viewed on the package GitHub page.
Clinical Informatics Dependencies
GENETEX facilitates abstraction of medical records for importation into the Genomics Instrument in REDCap®. The data dictionary for this form has been previously published3
Comparison of GENETEX to Manual Abstraction
Sample genomic reports were either generated or obtained from commercial vendors. These data were devoid of protected health information; thus, no IRB was required for this project. Two highly-trained abstractors manually abstracted the reports and recorded the time-to-capture for each report. GENETEX was used to abstract these same reports. To simulate the real-world experience, both techniques incorporated a manual visual quality-control step to verify if imported results were accurate. The time spent on this step was included in the total time-to-capture.. Agreement rates were compared using R. A paired wilcoxon test was used to compare the time of manual abstraction with the time to capture CGD with the GENETEX package.
Results
Inputs/User Interface
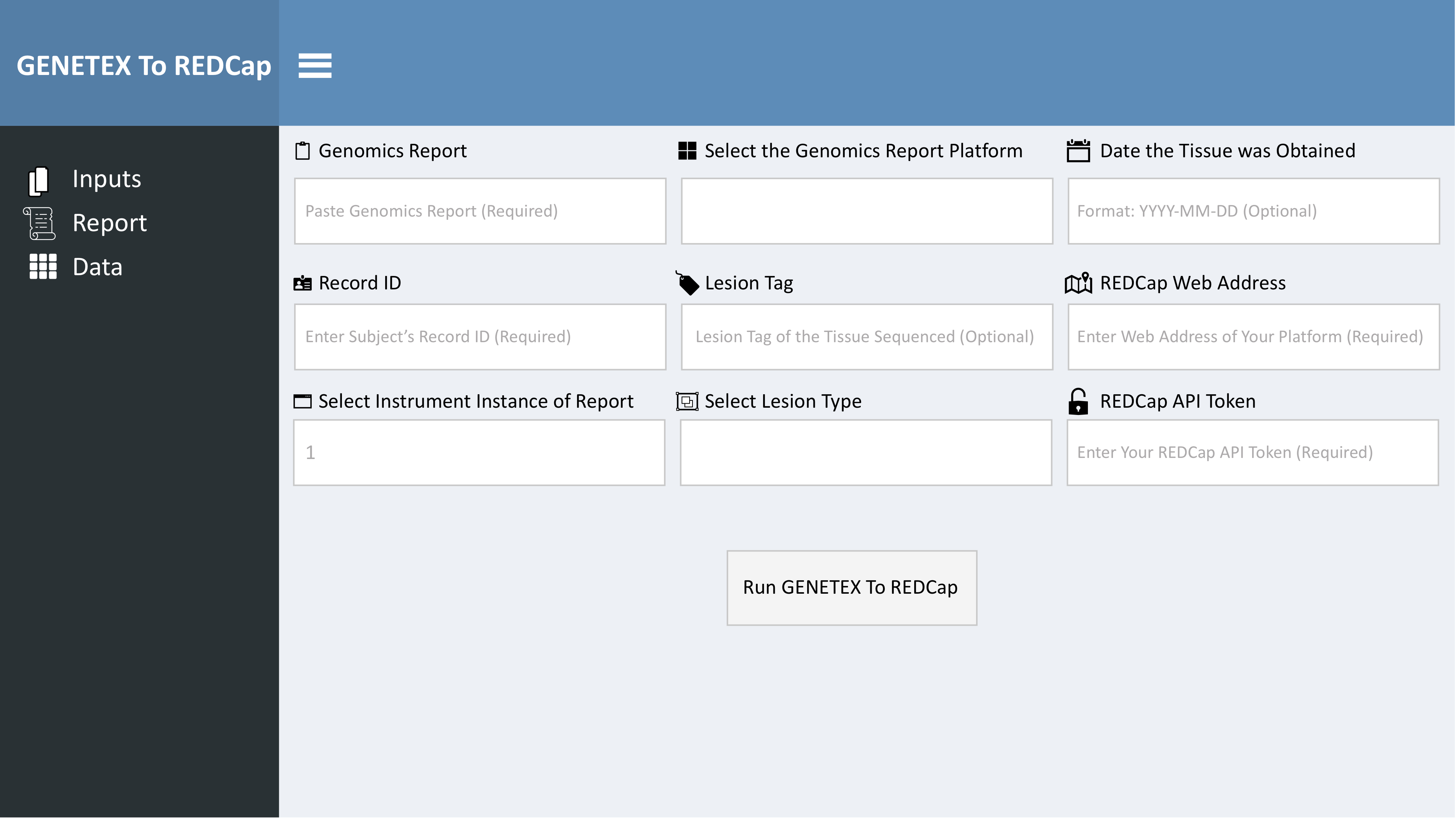
CGD in the real-world setting is predominantly contained in either portable document format (PDF) documents sent to providers by commercial vendors or in text files contained within EHR. Thus to facilitate abstraction of these data, we developed a browser-based user interface that incorporates text data captured on a clipboard as input in a Shiny application. Text is copied to a computer’s clipboard and pasted into the text area input in the Shiny application (Figure 2).
Figure 2: Browser-Based User Interface Depicted is the user interface (UI) of the Shiny app of GENETEX. This UI is produced by running the code in the R script “GENETEX Shiny app.R”, which can be found on GITHUB.
Users then control additional inputs including free text of the subject’s record id (required field), REDCap® instrument instance (required), lesion tag descriptor (optional field), and date the tissue was obtained (optional). Drop down inputs are also presented to the user including selection of the platform used to generate the genomics report (required) and the type of lesion the genomics report was generated from (e.g. primary lesion vs. metastases) (optional). Finally, to direct the data to REDCap®, users enter strings of the web address of the REDCap® platform (required) as well as the REDCap® API Token (required). These inputs are then called to the function genetex_to_redcap() by the action button “Run GENETEX to REDCap”.
Server Side Functions
The server side of the Shiny application contains the executable code of GENETEX. The package contains a set of functions that then parse, text mine and transform the input into structured data to serve as the substrate for import into REDCap® (Figure 1). Table 1 summarizes the functions and their respective function to extract key elements from the genomics report. We have methods to automatically mine HUGO Gene Nomenclature Committee-approved gene names and detected amino acid and/or nucleotide alterations, tumor mutational burden (tmb), mismatch repair status (mmr), copy number variants (cnv) and mutational signatures. In addition, our implementation transforms the data and links the appropriate CGD with the variables used in the Genomics Instrument so that they can be uploaded into REDCap®.
| Function | Functionality |
|---|---|
| genetex_to_redcap() | integrates key verbs to provide NLP tools to abstract data from a variety of genomic reports and import them to REDCap |
| gene.variants() | integrates various platform-specific NLP functions to text mine gene names and nucleotide variants from genomic reports and transforms them to structured data for import into REDCap |
| cnv() | integrates various platform-specific NLP functions to text mine gene names and copy number variants data from a variety of genomic reports and transforms them to structured data for import into REDCap |
| mmr() | text mines mismatch repair status from genomic reports and transforms it to structured data for import into REDCap |
| mutational.signatures() | text mines mutational signatures data from a variety of genomic reports and transforms it to structured data for import into REDCap |
| tmb() | text mines tumor mutation burden (TMB) data from a variety of genomic reports and transforms it to structured data for import into REDCap |
| platform() | applies regular expressions to assign a numerical value for the various platforms used for genomic reports that aligns with the `genomics_platform` field in the REDCap Genomics Instrument |
| genes_regex() | produces a regular expression of over 900 HGNC gene names |
| genes_boundary_regex() | produces a regular expression of over 900 HGNC gene names as a unique string with word boundaries |
| genomics.tissue.type() | applies regular expressions to assign a numerical value for the various platforms used for genomic reports that aligns with the `genomics_platform` field in the REDCap Genomics Instrument |
Table 1 - GENETEX Functions. Key functions unique to GENETEX with brief description of action are shown. Description of other functions can be found in the package’s Help Page.
Text Mining CGD
Overview of Data Processing
Following the initial step of securing CGD into the Shiny application, GENETEX converts these character strings to a data frame for text mining. At this time, CGD from the following platforms are able to be processed by GENETEX: Guardant360™15, FoundationOne®16, MGH SNaPshot17 and BWH Oncopanel18
Due to idiosyncratic differences between these reports, we developed platform-specific functions to text mine data. For example, gene.variants.isolate.oncopanel() and gene.variants.isolate.snapshot() isolate gene variant data from BWH Oncopanel and MGH SNaPshot reports, respectively. However, in general, following securing CGD, the data is tokenized using the cSplit() function from the splitstackshape package.
Isolating Genomic Data with Regular Expressions
To perform text mining of CGD, we created a number of regular expressions (regex) to identify gene names, nucleotide and amino acid sequences and cell-free DNA (cfDNA) data within genomic reports. For example, the function genes_boundary_regex() generates a regular expression of >900 gene names surrounded by word boundaries (Figure 3a). We further designed regular expressions to detect nucleotide and amino acid sequences and the magnitude of cfDNA (Figure 3b). The regular expressions were effectively combined to identify only those genomic information of interest (Figure 3C). Finally, we filter out unnecessary elements (e.g. the strings “RESULTS:” “Single” “nucleotide” “variants”) that are not intended to be captured in the Genomics instrument (Figure 3D)
genetex::genes_boundary_regex()# A tibble: 1 × 1
genes
<chr>
1 "\\bABCB1\\b|\\bABCB11\\b|\\bABCC3\\b|\\bABL1\\b|\\bABL2\\b|\\bACTA2\\b|\\bAC…Figure 3a: Regular expression of gene names. Depicted is a portion of the character vector output of the function genes_boundary_regex(). This function produces a regular expression that is used by GENETEX to identify gene names in CGD reports.
nuc_regex <- "[ACTG]>[ACTG]|del[ACTG]"
aa_regex <- "(\\b([A-Z][0-9]{1,}(([A-Z])|(_[A-Z][1-9]{1,}del)|(fs\\*[1-9]{1,})|(\\*)|(fs)|(del)))|(p\\.[A-Z]))|([0-9]ins[A-Z])"
cfdna_regex <- "\\b[0-9]{1,2}\\.[0-9]{1,2}%"Figure 3b: Regular expression of nucleotide and amino acid sequences, and cell-free DNA. Shown are the three regular expressions used to identify nucleotide sequences (“nuc_regex”), amino acid sequences (“aa_regex”) and cell-free DNA (“cfdna_regex”) contained within CGD reports.
| X |
|---|
| RESULTS: |
| Single |
| nucleotide |
| variants: |
| ATM |
| 2353C>T |
| R785C |
| 0.1% |
| PTEN |
| L257V |
| 769C>G |
| 8.1% |
Figure 3c: Tokenized genomics report. Depicted is a portion of a CGD report that has been tokenized. Here, each word of the report is partitioned into a single cell of the vector “X”.
genes_nuc_aa_cfdna_regex <- paste(genes_boundary_regex, aa_regex, nuc_regex, cfdna_regex, sep = "|")
dt.1 <- dt %>%
dplyr::filter(stringr::str_detect(string = dt$X,
pattern = stringr::regex(genes_nuc_aa_cfdna_regex)))| X |
|---|
| ATM |
| 2353C>T |
| R785C |
| 0.1% |
| PTEN |
| L257V |
| 769C>G |
| 8.1% |
Figure 3d: Example of report filtered with genes_nuc_aa_cfdna_regex. Shown is vector “X” from Figure 3c, which has been filtered by a regular expression that selects only cells with elements relevant to gene names, nucleotide and amino acid sequences and cell-free DNA. The code used for this step is demonstrated above the output.
Mapping REDCap® Variables
In addition to isolating key genomic data from reports, the above regular expressions are also used to map variables used in the Genomics Instrument. The instrument uses the following variable prefixes “variant_gene”, “variant_nucleotide”, “variant_protein” and “variant_gene_perc_cfdna” to enable tidy data for gene names, nucleotide variants, amino acid variants and percent cfDNA, respectively. Using the following combination of ifelse() statements, str_detect() and the aforementioned regular expressions, these variables can be linked to the corresponding tidy data (Figure 3f).
dt.3 <- dt.2 %>%
mutate(var = ifelse(test = str_detect(string = dt.2$X,
pattern = regex(genes_boundary_regex)),
yes = "variant_gene",
no = ifelse(test = str_detect(string = dt.2$X,
pattern = regex(nuc_regex)),
yes = "variant_nucleotide",
no = ifelse(test = str_detect(string = dt.2$X,
pattern = regex(aa_regex)),
yes = "variant_protein",
no = ifelse(test = str_detect(string = dt.2$X,
pattern = regex(cfdna_regex)),
yes = "variant_gene_perc_cfdna",
no = "")))))| X | keywords | group | var |
|---|---|---|---|
| ATM | TRUE | 1 | variant_gene |
| 2353C>T | FALSE | 1 | variant_nucleotide |
| R785C | FALSE | 1 | variant_protein |
| 0.1% | FALSE | 1 | variant_gene_perc_cfdna |
| PTEN | TRUE | 2 | variant_gene |
| L257V | FALSE | 2 | variant_protein |
| 769C>G | FALSE | 2 | variant_nucleotide |
| 8.1% | FALSE | 2 | variant_gene_perc_cfdna |
Figure 3f: Mapping REDCap variables to data elements. . Each tokenized data element in vector “X” must be linked with an appropriate variable name from the Genomics Instrument. In this step, the four relevant variable stems, “variant_gene”, “variant_nucleotide”, “variant_protein” and “variant_gene_perc_cfdna” are matched with the relevant data in vector “X” by combining the ifelse() and str_detect() functions with the regular expressions “genes_boundary_regex”, “nuc_regex”, “aa_regex” and “cfdna_regex”.
As previously described3, in order to produce unique variables that can be linked with other related information, the Genomics Instrument utilizes a Numeric Suffix Linker System (NSLS). The NSLS links related elements of CGD with a character string in the variable name (e.g. “variant”) with an underscore and a numeric (e.g “_1”). Therefore, a given gene will be grouped with its correlated nucleotide/amino acid variants and/or cfDNA information with a unique character string and numeric. For example, the variable prefixes “variant_gene”, “variant_nucleotide”, “variant_protein” and “variant_gene_perc_cfdna” will all be linked with the same numeric suffix (e.g. “_1”). Consequently, these elements can be grouped during analysis with the unique pairing of “variant” and “_1”. Therefore, a final step in creating this unique variable linked system involves pasting the “var” vector with the “group” vector(Figure 3g).
dt.4 <- dt.3 %>%
mutate(variables = paste(var, group, sep = "_"))| X | keywords | group | var | variables |
|---|---|---|---|---|
| ATM | TRUE | 1 | variant_gene | variant_gene_1 |
| 2353C>T | FALSE | 1 | variant_nucleotide | variant_nucleotide_1 |
| R785C | FALSE | 1 | variant_protein | variant_protein_1 |
| 0.1% | FALSE | 1 | variant_gene_perc_cfdna | variant_gene_perc_cfdna_1 |
| PTEN | TRUE | 2 | variant_gene | variant_gene_2 |
| L257V | FALSE | 2 | variant_protein | variant_protein_2 |
| 769C>G | FALSE | 2 | variant_nucleotide | variant_nucleotide_2 |
| 8.1% | FALSE | 2 | variant_gene_perc_cfdna | variant_gene_perc_cfdna_2 |
Figure 3g: Complete mapping REDCap variables to data elements with numeric suffix linker system. Each data element in vector “X” must correspond to a unique variable name to be imported into REDCap. Therefore, in this final step the variable stems “variant_gene”, “variant_nucleotide”, “variant_protein” and “variant_gene_perc_cfdna” and linked to the number found in the column “group” which produces a unique variable. All of those variables with the suffix “_1” will be “linked” together using a numeric suffix linker system.
An analogous approach is used to identify and abstract data on cnvs, tmb, mmr and mutational signatures. Please see the description file and annotated R scripts contained within the package in GitHub for further details.
Package Outputs
The front-end Shiny application is executed with an action button that produces two easy-to-view outputs. The first, which can be viewed by clicking on “Report” in the sidebar, produces a verbatimTextOutput of the genomic report to ensure that the correct report was pasted into the textAreaInput. The second one, viewed by clicking on “Data” in the sidebar, is a table of the data executed by genetex_to_redcap() (Supplemental Table 1). This is intended to provide the user with an output of the data generated by the function so that a quality-control step can take place.
| variables | results |
|---|---|
| record_id | Lewis Jones |
| redcap_repeat_instrument | genomics |
| redcap_repeat_instance | 1 |
| genomics_yn | 1 |
| lesion_tag_genomics | Liquid Biopsy |
| genomics_tissue_type | 5 |
| genomics_date | 2020-12-22 |
| genomics_platform | 6 |
| genomics_platform_other | |
| mmr | 2 |
| tmb_yn | 1 |
| tmb | 81.47 |
| tmb_abs | |
| mutation_signature_number | 0 |
| mutation_signature_1 | |
| mutation_signature_2 | |
| mutation_signature_3 | |
| mutation_signature_4 | |
| mutation_signature_5 | |
| mutation_signature_6 | |
| mutation_signature_7 | |
| mutation_signature_8 | |
| mutation_signature_9 | |
| mutation_signature_10 | |
| variant_number | 20 |
| variant_gene_1 | NTRK2 |
| variant_nucleotide_1 | |
| variant_protein_1 | A31T |
| variant_gene_perc_cfdna_1 | 3.5% |
| variant_gene_2 | CTNNB1 |
| variant_nucleotide_2 | |
| variant_protein_2 | R550R |
| variant_gene_perc_cfdna_2 | 3.2% |
| variant_gene_3 | PTEN |
| variant_nucleotide_3 | |
| variant_protein_3 | E157fs |
| variant_gene_perc_cfdna_3 | 3.2% |
| variant_gene_4 | PIK3CA |
| variant_nucleotide_4 | |
| variant_protein_4 | E545K |
| variant_gene_perc_cfdna_4 | 2.6% |
| variant_gene_5 | FANCA |
| variant_nucleotide_5 | |
| variant_protein_5 | R1400H |
| variant_gene_perc_cfdna_5 | 2.5% |
| variant_gene_6 | PTEN |
| variant_nucleotide_6 | |
| variant_protein_6 | K267fs |
| variant_gene_perc_cfdna_6 | 2.5% |
| variant_gene_7 | BRCA2 |
| variant_nucleotide_7 | |
| variant_protein_7 | I605fs |
| variant_gene_perc_cfdna_7 | 2.3% |
| variant_gene_8 | BRAF |
| variant_nucleotide_8 | |
| variant_protein_8 | A762V |
| variant_gene_perc_cfdna_8 | 2.2% |
| variant_gene_9 | PALB2 |
| variant_nucleotide_9 | |
| variant_protein_9 | R37C |
| variant_gene_perc_cfdna_9 | 1.9% |
| variant_gene_10 | CHEK2 |
| variant_nucleotide_10 | |
| variant_protein_10 | R346H |
| variant_gene_perc_cfdna_10 | 1.8% |
| variant_gene_11 | NTRK1 |
| variant_nucleotide_11 | |
| variant_protein_11 | T741T |
| variant_gene_perc_cfdna_11 | 1.7% |
| variant_gene_12 | AR |
| variant_nucleotide_12 | |
| variant_protein_12 | H875Y |
| variant_gene_perc_cfdna_12 | 0.5% |
| variant_gene_13 | MTOR |
| variant_nucleotide_13 | |
| variant_protein_13 | W1456R |
| variant_gene_perc_cfdna_13 | 0.4% |
| variant_gene_14 | NOTCH1 |
| variant_nucleotide_14 | |
| variant_protein_14 | F357del |
| variant_gene_perc_cfdna_14 | 0.4% |
| variant_gene_15 | DDR2 |
| variant_nucleotide_15 | |
| variant_protein_15 | P157L |
| variant_gene_perc_cfdna_15 | 0.3% |
| variant_gene_16 | GNA11 |
| variant_nucleotide_16 | |
| variant_protein_16 | G208fs |
| variant_gene_perc_cfdna_16 | 0.3% |
| variant_gene_17 | MPL |
| variant_nucleotide_17 | |
| variant_protein_17 | P530P |
| variant_gene_perc_cfdna_17 | 0.3% |
| variant_gene_18 | ALK |
| variant_nucleotide_18 | |
| variant_protein_18 | Y1584Y |
| variant_gene_perc_cfdna_18 | 0.2% |
| variant_gene_19 | NOTCH1 |
| variant_nucleotide_19 | |
| variant_protein_19 | S2486fs |
| variant_gene_perc_cfdna_19 | 0.2% |
| variant_gene_20 | ARID1A |
| variant_nucleotide_20 | |
| variant_protein_20 | P1710P |
| variant_gene_perc_cfdna_20 | 0.1% |
| variant_gene_21 | |
| variant_nucleotide_21 | |
| variant_protein_21 | |
| variant_gene_perc_cfdna_21 | |
| variant_gene_22 | |
| variant_nucleotide_22 | |
| variant_protein_22 | |
| variant_gene_perc_cfdna_22 | |
| variant_gene_23 | |
| variant_nucleotide_23 | |
| variant_protein_23 | |
| variant_gene_perc_cfdna_23 | |
| variant_gene_24 | |
| variant_nucleotide_24 | |
| variant_protein_24 | |
| variant_gene_perc_cfdna_24 | |
| variant_gene_25 | |
| variant_nucleotide_25 | |
| variant_protein_25 | |
| variant_gene_perc_cfdna_25 | |
| variant_gene_26 | |
| variant_nucleotide_26 | |
| variant_protein_26 | |
| variant_gene_perc_cfdna_26 | |
| variant_gene_27 | |
| variant_nucleotide_27 | |
| variant_protein_27 | |
| variant_gene_perc_cfdna_27 | |
| variant_gene_28 | |
| variant_nucleotide_28 | |
| variant_protein_28 | |
| variant_gene_perc_cfdna_28 | |
| variant_gene_29 | |
| variant_nucleotide_29 | |
| variant_protein_29 | |
| variant_gene_perc_cfdna_29 | |
| variant_gene_30 | |
| variant_nucleotide_30 | |
| variant_protein_30 | |
| variant_gene_perc_cfdna_30 | |
| variant_gene_31 | |
| variant_nucleotide_31 | |
| variant_protein_31 | |
| variant_gene_perc_cfdna_31 | |
| variant_gene_32 | |
| variant_nucleotide_32 | |
| variant_protein_32 | |
| variant_gene_perc_cfdna_32 | |
| variant_gene_33 | |
| variant_nucleotide_33 | |
| variant_protein_33 | |
| variant_gene_perc_cfdna_33 | |
| variant_gene_34 | |
| variant_nucleotide_34 | |
| variant_protein_34 | |
| variant_gene_perc_cfdna_34 | |
| variant_gene_35 | |
| variant_nucleotide_35 | |
| variant_protein_35 | |
| variant_gene_perc_cfdna_35 | |
| variant_gene_36 | |
| variant_nucleotide_36 | |
| variant_protein_36 | |
| variant_gene_perc_cfdna_36 | |
| variant_gene_37 | |
| variant_nucleotide_37 | |
| variant_protein_37 | |
| variant_gene_perc_cfdna_37 | |
| variant_gene_38 | |
| variant_nucleotide_38 | |
| variant_protein_38 | |
| variant_gene_perc_cfdna_38 | |
| variant_gene_39 | |
| variant_nucleotide_39 | |
| variant_protein_39 | |
| variant_gene_perc_cfdna_39 | |
| variant_gene_40 | |
| variant_nucleotide_40 | |
| variant_protein_40 | |
| variant_gene_perc_cfdna_40 | |
| variant_gene_41 | |
| variant_nucleotide_41 | |
| variant_protein_41 | |
| variant_gene_perc_cfdna_41 | |
| variant_gene_42 | |
| variant_nucleotide_42 | |
| variant_protein_42 | |
| variant_gene_perc_cfdna_42 | |
| variant_gene_43 | |
| variant_nucleotide_43 | |
| variant_protein_43 | |
| variant_gene_perc_cfdna_43 | |
| variant_gene_44 | |
| variant_nucleotide_44 | |
| variant_protein_44 | |
| variant_gene_perc_cfdna_44 | |
| variant_gene_45 | |
| variant_nucleotide_45 | |
| variant_protein_45 | |
| variant_gene_perc_cfdna_45 | |
| variant_gene_46 | |
| variant_nucleotide_46 | |
| variant_protein_46 | |
| variant_gene_perc_cfdna_46 | |
| variant_gene_47 | |
| variant_nucleotide_47 | |
| variant_protein_47 | |
| variant_gene_perc_cfdna_47 | |
| variant_gene_48 | |
| variant_nucleotide_48 | |
| variant_protein_48 | |
| variant_gene_perc_cfdna_48 | |
| variant_gene_49 | |
| variant_nucleotide_49 | |
| variant_protein_49 | |
| variant_gene_perc_cfdna_49 | |
| variant_gene_50 | |
| variant_nucleotide_50 | |
| variant_protein_50 | |
| variant_gene_perc_cfdna_50 | |
| variant_gene_51 | |
| variant_nucleotide_51 | |
| variant_protein_51 | |
| variant_gene_perc_cfdna_51 | |
| variant_gene_52 | |
| variant_nucleotide_52 | |
| variant_protein_52 | |
| variant_gene_perc_cfdna_52 | |
| variant_gene_53 | |
| variant_nucleotide_53 | |
| variant_protein_53 | |
| variant_gene_perc_cfdna_53 | |
| variant_gene_54 | |
| variant_nucleotide_54 | |
| variant_protein_54 | |
| variant_gene_perc_cfdna_54 | |
| variant_gene_55 | |
| variant_nucleotide_55 | |
| variant_protein_55 | |
| variant_gene_perc_cfdna_55 | |
| variant_gene_56 | |
| variant_nucleotide_56 | |
| variant_protein_56 | |
| variant_gene_perc_cfdna_56 | |
| variant_gene_57 | |
| variant_nucleotide_57 | |
| variant_protein_57 | |
| variant_gene_perc_cfdna_57 | |
| variant_gene_58 | |
| variant_nucleotide_58 | |
| variant_protein_58 | |
| variant_gene_perc_cfdna_58 | |
| variant_gene_59 | |
| variant_nucleotide_59 | |
| variant_protein_59 | |
| variant_gene_perc_cfdna_59 | |
| variant_gene_60 | |
| variant_nucleotide_60 | |
| variant_protein_60 | |
| variant_gene_perc_cfdna_60 | |
| variant_gene_61 | |
| variant_nucleotide_61 | |
| variant_protein_61 | |
| variant_gene_perc_cfdna_61 | |
| variant_gene_62 | |
| variant_nucleotide_62 | |
| variant_protein_62 | |
| variant_gene_perc_cfdna_62 | |
| variant_gene_63 | |
| variant_nucleotide_63 | |
| variant_protein_63 | |
| variant_gene_perc_cfdna_63 | |
| variant_gene_64 | |
| variant_nucleotide_64 | |
| variant_protein_64 | |
| variant_gene_perc_cfdna_64 | |
| variant_gene_65 | |
| variant_nucleotide_65 | |
| variant_protein_65 | |
| variant_gene_perc_cfdna_65 | |
| variant_gene_66 | |
| variant_nucleotide_66 | |
| variant_protein_66 | |
| variant_gene_perc_cfdna_66 | |
| variant_gene_67 | |
| variant_nucleotide_67 | |
| variant_protein_67 | |
| variant_gene_perc_cfdna_67 | |
| variant_gene_68 | |
| variant_nucleotide_68 | |
| variant_protein_68 | |
| variant_gene_perc_cfdna_68 | |
| variant_gene_69 | |
| variant_nucleotide_69 | |
| variant_protein_69 | |
| variant_gene_perc_cfdna_69 | |
| variant_gene_70 | |
| variant_nucleotide_70 | |
| variant_protein_70 | |
| variant_gene_perc_cfdna_70 | |
| variant_gene_71 | |
| variant_nucleotide_71 | |
| variant_protein_71 | |
| variant_gene_perc_cfdna_71 | |
| variant_gene_72 | |
| variant_nucleotide_72 | |
| variant_protein_72 | |
| variant_gene_perc_cfdna_72 | |
| variant_gene_73 | |
| variant_nucleotide_73 | |
| variant_protein_73 | |
| variant_gene_perc_cfdna_73 | |
| variant_gene_74 | |
| variant_nucleotide_74 | |
| variant_protein_74 | |
| variant_gene_perc_cfdna_74 | |
| variant_gene_75 | |
| variant_nucleotide_75 | |
| variant_protein_75 | |
| variant_gene_perc_cfdna_75 | |
| variant_gene_76 | |
| variant_nucleotide_76 | |
| variant_protein_76 | |
| variant_gene_perc_cfdna_76 | |
| variant_gene_77 | |
| variant_nucleotide_77 | |
| variant_protein_77 | |
| variant_gene_perc_cfdna_77 | |
| variant_gene_78 | |
| variant_nucleotide_78 | |
| variant_protein_78 | |
| variant_gene_perc_cfdna_78 | |
| variant_gene_79 | |
| variant_nucleotide_79 | |
| variant_protein_79 | |
| variant_gene_perc_cfdna_79 | |
| variant_gene_80 | |
| variant_nucleotide_80 | |
| variant_protein_80 | |
| variant_gene_perc_cfdna_80 | |
| variant_gene_81 | |
| variant_nucleotide_81 | |
| variant_protein_81 | |
| variant_gene_perc_cfdna_81 | |
| variant_gene_82 | |
| variant_nucleotide_82 | |
| variant_protein_82 | |
| variant_gene_perc_cfdna_82 | |
| variant_gene_83 | |
| variant_nucleotide_83 | |
| variant_protein_83 | |
| variant_gene_perc_cfdna_83 | |
| variant_gene_84 | |
| variant_nucleotide_84 | |
| variant_protein_84 | |
| variant_gene_perc_cfdna_84 | |
| variant_gene_85 | |
| variant_nucleotide_85 | |
| variant_protein_85 | |
| variant_gene_perc_cfdna_85 | |
| variant_gene_86 | |
| variant_nucleotide_86 | |
| variant_protein_86 | |
| variant_gene_perc_cfdna_86 | |
| variant_gene_87 | |
| variant_nucleotide_87 | |
| variant_protein_87 | |
| variant_gene_perc_cfdna_87 | |
| variant_gene_88 | |
| variant_nucleotide_88 | |
| variant_protein_88 | |
| variant_gene_perc_cfdna_88 | |
| variant_gene_89 | |
| variant_nucleotide_89 | |
| variant_protein_89 | |
| variant_gene_perc_cfdna_89 | |
| variant_gene_90 | |
| variant_nucleotide_90 | |
| variant_protein_90 | |
| variant_gene_perc_cfdna_90 | |
| variant_gene_91 | |
| variant_nucleotide_91 | |
| variant_protein_91 | |
| variant_gene_perc_cfdna_91 | |
| variant_gene_92 | |
| variant_nucleotide_92 | |
| variant_protein_92 | |
| variant_gene_perc_cfdna_92 | |
| variant_gene_93 | |
| variant_nucleotide_93 | |
| variant_protein_93 | |
| variant_gene_perc_cfdna_93 | |
| variant_gene_94 | |
| variant_nucleotide_94 | |
| variant_protein_94 | |
| variant_gene_perc_cfdna_94 | |
| variant_gene_95 | |
| variant_nucleotide_95 | |
| variant_protein_95 | |
| variant_gene_perc_cfdna_95 | |
| variant_gene_96 | |
| variant_nucleotide_96 | |
| variant_protein_96 | |
| variant_gene_perc_cfdna_96 | |
| variant_gene_97 | |
| variant_nucleotide_97 | |
| variant_protein_97 | |
| variant_gene_perc_cfdna_97 | |
| variant_gene_98 | |
| variant_nucleotide_98 | |
| variant_protein_98 | |
| variant_gene_perc_cfdna_98 | |
| variant_gene_99 | |
| variant_nucleotide_99 | |
| variant_protein_99 | |
| variant_gene_perc_cfdna_99 | |
| variant_gene_100 | |
| variant_nucleotide_100 | |
| variant_protein_100 | |
| variant_gene_perc_cfdna_100 | |
| variant_gene_101 | |
| variant_nucleotide_101 | |
| variant_protein_101 | |
| variant_gene_perc_cfdna_101 | |
| variant_gene_102 | |
| variant_nucleotide_102 | |
| variant_protein_102 | |
| variant_gene_perc_cfdna_102 | |
| variant_gene_103 | |
| variant_nucleotide_103 | |
| variant_protein_103 | |
| variant_gene_perc_cfdna_103 | |
| variant_gene_104 | |
| variant_nucleotide_104 | |
| variant_protein_104 | |
| variant_gene_perc_cfdna_104 | |
| variant_gene_105 | |
| variant_nucleotide_105 | |
| variant_protein_105 | |
| variant_gene_perc_cfdna_105 | |
| variant_gene_106 | |
| variant_nucleotide_106 | |
| variant_protein_106 | |
| variant_gene_perc_cfdna_106 | |
| variant_gene_107 | |
| variant_nucleotide_107 | |
| variant_protein_107 | |
| variant_gene_perc_cfdna_107 | |
| variant_gene_108 | |
| variant_nucleotide_108 | |
| variant_protein_108 | |
| variant_gene_perc_cfdna_108 | |
| variant_gene_109 | |
| variant_nucleotide_109 | |
| variant_protein_109 | |
| variant_gene_perc_cfdna_109 | |
| variant_gene_110 | |
| variant_nucleotide_110 | |
| variant_protein_110 | |
| variant_gene_perc_cfdna_110 | |
| variant_gene_111 | |
| variant_nucleotide_111 | |
| variant_protein_111 | |
| variant_gene_perc_cfdna_111 | |
| variant_gene_112 | |
| variant_nucleotide_112 | |
| variant_protein_112 | |
| variant_gene_perc_cfdna_112 | |
| variant_gene_113 | |
| variant_nucleotide_113 | |
| variant_protein_113 | |
| variant_gene_perc_cfdna_113 | |
| variant_gene_114 | |
| variant_nucleotide_114 | |
| variant_protein_114 | |
| variant_gene_perc_cfdna_114 | |
| variant_gene_115 | |
| variant_nucleotide_115 | |
| variant_protein_115 | |
| variant_gene_perc_cfdna_115 | |
| variant_gene_116 | |
| variant_nucleotide_116 | |
| variant_protein_116 | |
| variant_gene_perc_cfdna_116 | |
| variant_gene_117 | |
| variant_nucleotide_117 | |
| variant_protein_117 | |
| variant_gene_perc_cfdna_117 | |
| variant_gene_118 | |
| variant_nucleotide_118 | |
| variant_protein_118 | |
| variant_gene_perc_cfdna_118 | |
| variant_gene_119 | |
| variant_nucleotide_119 | |
| variant_protein_119 | |
| variant_gene_perc_cfdna_119 | |
| variant_gene_120 | |
| variant_nucleotide_120 | |
| variant_protein_120 | |
| variant_gene_perc_cfdna_120 | |
| variant_gene_121 | |
| variant_nucleotide_121 | |
| variant_protein_121 | |
| variant_gene_perc_cfdna_121 | |
| variant_gene_122 | |
| variant_nucleotide_122 | |
| variant_protein_122 | |
| variant_gene_perc_cfdna_122 | |
| variant_gene_123 | |
| variant_nucleotide_123 | |
| variant_protein_123 | |
| variant_gene_perc_cfdna_123 | |
| variant_gene_124 | |
| variant_nucleotide_124 | |
| variant_protein_124 | |
| variant_gene_perc_cfdna_124 | |
| variant_gene_125 | |
| variant_nucleotide_125 | |
| variant_protein_125 | |
| variant_gene_perc_cfdna_125 | |
| variant_gene_126 | |
| variant_nucleotide_126 | |
| variant_protein_126 | |
| variant_gene_perc_cfdna_126 | |
| variant_gene_127 | |
| variant_nucleotide_127 | |
| variant_protein_127 | |
| variant_gene_perc_cfdna_127 | |
| variant_gene_128 | |
| variant_nucleotide_128 | |
| variant_protein_128 | |
| variant_gene_perc_cfdna_128 | |
| variant_gene_129 | |
| variant_nucleotide_129 | |
| variant_protein_129 | |
| variant_gene_perc_cfdna_129 | |
| variant_gene_130 | |
| variant_nucleotide_130 | |
| variant_protein_130 | |
| variant_gene_perc_cfdna_130 | |
| variant_gene_131 | |
| variant_nucleotide_131 | |
| variant_protein_131 | |
| variant_gene_perc_cfdna_131 | |
| variant_gene_132 | |
| variant_nucleotide_132 | |
| variant_protein_132 | |
| variant_gene_perc_cfdna_132 | |
| variant_gene_133 | |
| variant_nucleotide_133 | |
| variant_protein_133 | |
| variant_gene_perc_cfdna_133 | |
| variant_gene_134 | |
| variant_nucleotide_134 | |
| variant_protein_134 | |
| variant_gene_perc_cfdna_134 | |
| variant_gene_135 | |
| variant_nucleotide_135 | |
| variant_protein_135 | |
| variant_gene_perc_cfdna_135 | |
| variant_gene_136 | |
| variant_nucleotide_136 | |
| variant_protein_136 | |
| variant_gene_perc_cfdna_136 | |
| variant_gene_137 | |
| variant_nucleotide_137 | |
| variant_protein_137 | |
| variant_gene_perc_cfdna_137 | |
| variant_gene_138 | |
| variant_nucleotide_138 | |
| variant_protein_138 | |
| variant_gene_perc_cfdna_138 | |
| variant_gene_139 | |
| variant_nucleotide_139 | |
| variant_protein_139 | |
| variant_gene_perc_cfdna_139 | |
| variant_gene_140 | |
| variant_nucleotide_140 | |
| variant_protein_140 | |
| variant_gene_perc_cfdna_140 | |
| variant_gene_141 | |
| variant_nucleotide_141 | |
| variant_protein_141 | |
| variant_gene_perc_cfdna_141 | |
| variant_gene_142 | |
| variant_nucleotide_142 | |
| variant_protein_142 | |
| variant_gene_perc_cfdna_142 | |
| variant_gene_143 | |
| variant_nucleotide_143 | |
| variant_protein_143 | |
| variant_gene_perc_cfdna_143 | |
| variant_gene_144 | |
| variant_nucleotide_144 | |
| variant_protein_144 | |
| variant_gene_perc_cfdna_144 | |
| variant_gene_145 | |
| variant_nucleotide_145 | |
| variant_protein_145 | |
| variant_gene_perc_cfdna_145 | |
| variant_gene_146 | |
| variant_nucleotide_146 | |
| variant_protein_146 | |
| variant_gene_perc_cfdna_146 | |
| variant_gene_147 | |
| variant_nucleotide_147 | |
| variant_protein_147 | |
| variant_gene_perc_cfdna_147 | |
| variant_gene_148 | |
| variant_nucleotide_148 | |
| variant_protein_148 | |
| variant_gene_perc_cfdna_148 | |
| variant_gene_149 | |
| variant_nucleotide_149 | |
| variant_protein_149 | |
| variant_gene_perc_cfdna_149 | |
| variant_gene_150 | |
| variant_nucleotide_150 | |
| variant_protein_150 | |
| variant_gene_perc_cfdna_150 | |
| variant_gene_151 | |
| variant_nucleotide_151 | |
| variant_protein_151 | |
| variant_gene_152 | |
| variant_nucleotide_152 | |
| variant_protein_152 | |
| variant_gene_153 | |
| variant_nucleotide_153 | |
| variant_protein_153 | |
| variant_gene_154 | |
| variant_nucleotide_154 | |
| variant_protein_154 | |
| variant_gene_155 | |
| variant_nucleotide_155 | |
| variant_protein_155 | |
| variant_gene_156 | |
| variant_nucleotide_156 | |
| variant_protein_156 | |
| variant_gene_157 | |
| variant_nucleotide_157 | |
| variant_protein_157 | |
| variant_gene_158 | |
| variant_nucleotide_158 | |
| variant_protein_158 | |
| variant_gene_159 | |
| variant_nucleotide_159 | |
| variant_protein_159 | |
| variant_gene_160 | |
| variant_nucleotide_160 | |
| variant_protein_160 | |
| variant_gene_161 | |
| variant_nucleotide_161 | |
| variant_protein_161 | |
| variant_gene_162 | |
| variant_nucleotide_162 | |
| variant_protein_162 | |
| variant_gene_163 | |
| variant_nucleotide_163 | |
| variant_protein_163 | |
| variant_gene_164 | |
| variant_nucleotide_164 | |
| variant_protein_164 | |
| variant_gene_165 | |
| variant_nucleotide_165 | |
| variant_protein_165 | |
| variant_gene_166 | |
| variant_nucleotide_166 | |
| variant_protein_166 | |
| variant_gene_167 | |
| variant_nucleotide_167 | |
| variant_protein_167 | |
| variant_gene_168 | |
| variant_nucleotide_168 | |
| variant_protein_168 | |
| variant_gene_169 | |
| variant_nucleotide_169 | |
| variant_protein_169 | |
| variant_gene_170 | |
| variant_nucleotide_170 | |
| variant_protein_170 | |
| variant_gene_171 | |
| variant_nucleotide_171 | |
| variant_protein_171 | |
| variant_gene_172 | |
| variant_nucleotide_172 | |
| variant_protein_172 | |
| variant_gene_173 | |
| variant_nucleotide_173 | |
| variant_protein_173 | |
| variant_gene_174 | |
| variant_nucleotide_174 | |
| variant_protein_174 | |
| variant_gene_175 | |
| variant_nucleotide_175 | |
| variant_protein_175 | |
| variant_gene_176 | |
| variant_nucleotide_176 | |
| variant_protein_176 | |
| variant_gene_177 | |
| variant_nucleotide_177 | |
| variant_protein_177 | |
| variant_gene_178 | |
| variant_nucleotide_178 | |
| variant_protein_178 | |
| variant_gene_179 | |
| variant_nucleotide_179 | |
| variant_protein_179 | |
| variant_gene_180 | |
| variant_nucleotide_180 | |
| variant_protein_180 | |
| variant_gene_181 | |
| variant_nucleotide_181 | |
| variant_protein_181 | |
| variant_gene_182 | |
| variant_nucleotide_182 | |
| variant_protein_182 | |
| variant_gene_183 | |
| variant_nucleotide_183 | |
| variant_protein_183 | |
| variant_gene_184 | |
| variant_nucleotide_184 | |
| variant_protein_184 | |
| variant_gene_185 | |
| variant_nucleotide_185 | |
| variant_protein_185 | |
| variant_gene_186 | |
| variant_nucleotide_186 | |
| variant_protein_186 | |
| variant_gene_187 | |
| variant_nucleotide_187 | |
| variant_protein_187 | |
| variant_gene_188 | |
| variant_nucleotide_188 | |
| variant_protein_188 | |
| variant_gene_189 | |
| variant_nucleotide_189 | |
| variant_protein_189 | |
| variant_gene_190 | |
| variant_nucleotide_190 | |
| variant_protein_190 | |
| variant_gene_191 | |
| variant_nucleotide_191 | |
| variant_protein_191 | |
| variant_gene_192 | |
| variant_nucleotide_192 | |
| variant_protein_192 | |
| variant_gene_193 | |
| variant_nucleotide_193 | |
| variant_protein_193 | |
| variant_gene_194 | |
| variant_nucleotide_194 | |
| variant_protein_194 | |
| variant_gene_195 | |
| variant_nucleotide_195 | |
| variant_protein_195 | |
| variant_gene_196 | |
| variant_nucleotide_196 | |
| variant_protein_196 | |
| variant_gene_197 | |
| variant_nucleotide_197 | |
| variant_protein_197 | |
| variant_gene_198 | |
| variant_nucleotide_198 | |
| variant_protein_198 | |
| variant_gene_199 | |
| variant_nucleotide_199 | |
| variant_protein_199 | |
| variant_gene_200 | |
| variant_nucleotide_200 | |
| variant_protein_200 | |
| variant_gene_201 | |
| variant_nucleotide_201 | |
| variant_protein_201 | |
| variant_gene_202 | |
| variant_nucleotide_202 | |
| variant_protein_202 | |
| variant_gene_203 | |
| variant_nucleotide_203 | |
| variant_protein_203 | |
| variant_gene_204 | |
| variant_nucleotide_204 | |
| variant_protein_204 | |
| variant_gene_205 | |
| variant_nucleotide_205 | |
| variant_protein_205 | |
| variant_gene_206 | |
| variant_nucleotide_206 | |
| variant_protein_206 | |
| variant_gene_207 | |
| variant_nucleotide_207 | |
| variant_protein_207 | |
| variant_gene_208 | |
| variant_nucleotide_208 | |
| variant_protein_208 | |
| variant_gene_209 | |
| variant_nucleotide_209 | |
| variant_protein_209 | |
| variant_gene_210 | |
| variant_nucleotide_210 | |
| variant_protein_210 | |
| variant_gene_211 | |
| variant_nucleotide_211 | |
| variant_protein_211 | |
| variant_gene_212 | |
| variant_nucleotide_212 | |
| variant_protein_212 | |
| variant_gene_213 | |
| variant_nucleotide_213 | |
| variant_protein_213 | |
| variant_gene_214 | |
| variant_nucleotide_214 | |
| variant_protein_214 | |
| variant_gene_215 | |
| variant_nucleotide_215 | |
| variant_protein_215 | |
| variant_gene_216 | |
| variant_nucleotide_216 | |
| variant_protein_216 | |
| variant_gene_217 | |
| variant_nucleotide_217 | |
| variant_protein_217 | |
| variant_gene_218 | |
| variant_nucleotide_218 | |
| variant_protein_218 | |
| variant_gene_219 | |
| variant_nucleotide_219 | |
| variant_protein_219 | |
| variant_gene_220 | |
| variant_nucleotide_220 | |
| variant_protein_220 | |
| variant_gene_221 | |
| variant_nucleotide_221 | |
| variant_protein_221 | |
| variant_gene_222 | |
| variant_nucleotide_222 | |
| variant_protein_222 | |
| variant_gene_223 | |
| variant_nucleotide_223 | |
| variant_protein_223 | |
| variant_gene_224 | |
| variant_nucleotide_224 | |
| variant_protein_224 | |
| variant_gene_225 | |
| variant_nucleotide_225 | |
| variant_protein_225 | |
| variant_gene_226 | |
| variant_nucleotide_226 | |
| variant_protein_226 | |
| variant_gene_227 | |
| variant_nucleotide_227 | |
| variant_protein_227 | |
| variant_gene_228 | |
| variant_nucleotide_228 | |
| variant_protein_228 | |
| variant_gene_229 | |
| variant_nucleotide_229 | |
| variant_protein_229 | |
| variant_gene_230 | |
| variant_nucleotide_230 | |
| variant_protein_230 | |
| variant_gene_231 | |
| variant_nucleotide_231 | |
| variant_protein_231 | |
| variant_gene_232 | |
| variant_nucleotide_232 | |
| variant_protein_232 | |
| variant_gene_233 | |
| variant_nucleotide_233 | |
| variant_protein_233 | |
| variant_gene_234 | |
| variant_nucleotide_234 | |
| variant_protein_234 | |
| variant_gene_235 | |
| variant_nucleotide_235 | |
| variant_protein_235 | |
| variant_gene_236 | |
| variant_nucleotide_236 | |
| variant_protein_236 | |
| variant_gene_237 | |
| variant_nucleotide_237 | |
| variant_protein_237 | |
| variant_gene_238 | |
| variant_nucleotide_238 | |
| variant_protein_238 | |
| variant_gene_239 | |
| variant_nucleotide_239 | |
| variant_protein_239 | |
| variant_gene_240 | |
| variant_nucleotide_240 | |
| variant_protein_240 | |
| variant_gene_241 | |
| variant_nucleotide_241 | |
| variant_protein_241 | |
| variant_gene_242 | |
| variant_nucleotide_242 | |
| variant_protein_242 | |
| variant_gene_243 | |
| variant_nucleotide_243 | |
| variant_protein_243 | |
| variant_gene_244 | |
| variant_nucleotide_244 | |
| variant_protein_244 | |
| variant_gene_245 | |
| variant_nucleotide_245 | |
| variant_protein_245 | |
| variant_gene_246 | |
| variant_nucleotide_246 | |
| variant_protein_246 | |
| variant_gene_247 | |
| variant_nucleotide_247 | |
| variant_protein_247 | |
| variant_gene_248 | |
| variant_nucleotide_248 | |
| variant_protein_248 | |
| variant_gene_249 | |
| variant_nucleotide_249 | |
| variant_protein_249 | |
| variant_gene_250 | |
| variant_nucleotide_250 | |
| variant_protein_250 | |
| variant_gene_251 | |
| variant_nucleotide_251 | |
| variant_protein_251 | |
| variant_gene_252 | |
| variant_nucleotide_252 | |
| variant_protein_252 | |
| variant_gene_253 | |
| variant_nucleotide_253 | |
| variant_protein_253 | |
| variant_gene_254 | |
| variant_nucleotide_254 | |
| variant_protein_254 | |
| variant_gene_255 | |
| variant_nucleotide_255 | |
| variant_protein_255 | |
| variant_gene_256 | |
| variant_nucleotide_256 | |
| variant_protein_256 | |
| variant_gene_257 | |
| variant_nucleotide_257 | |
| variant_protein_257 | |
| variant_gene_258 | |
| variant_nucleotide_258 | |
| variant_protein_258 | |
| variant_gene_259 | |
| variant_nucleotide_259 | |
| variant_protein_259 | |
| variant_gene_260 | |
| variant_nucleotide_260 | |
| variant_protein_260 | |
| variant_gene_261 | |
| variant_nucleotide_261 | |
| variant_protein_261 | |
| variant_gene_262 | |
| variant_nucleotide_262 | |
| variant_protein_262 | |
| variant_gene_263 | |
| variant_nucleotide_263 | |
| variant_protein_263 | |
| variant_gene_264 | |
| variant_nucleotide_264 | |
| variant_protein_264 | |
| variant_gene_265 | |
| variant_nucleotide_265 | |
| variant_protein_265 | |
| variant_gene_266 | |
| variant_nucleotide_266 | |
| variant_protein_266 | |
| variant_gene_267 | |
| variant_nucleotide_267 | |
| variant_protein_267 | |
| variant_gene_268 | |
| variant_nucleotide_268 | |
| variant_protein_268 | |
| variant_gene_269 | |
| variant_nucleotide_269 | |
| variant_protein_269 | |
| variant_gene_270 | |
| variant_nucleotide_270 | |
| variant_protein_270 | |
| variant_gene_271 | |
| variant_nucleotide_271 | |
| variant_protein_271 | |
| variant_gene_272 | |
| variant_nucleotide_272 | |
| variant_protein_272 | |
| variant_gene_273 | |
| variant_nucleotide_273 | |
| variant_protein_273 | |
| variant_gene_274 | |
| variant_nucleotide_274 | |
| variant_protein_274 | |
| variant_gene_275 | |
| variant_nucleotide_275 | |
| variant_protein_275 | |
| variant_gene_276 | |
| variant_nucleotide_276 | |
| variant_protein_276 | |
| variant_gene_277 | |
| variant_nucleotide_277 | |
| variant_protein_277 | |
| variant_gene_278 | |
| variant_nucleotide_278 | |
| variant_protein_278 | |
| variant_gene_279 | |
| variant_nucleotide_279 | |
| variant_protein_279 | |
| variant_gene_280 | |
| variant_nucleotide_280 | |
| variant_protein_280 | |
| variant_gene_281 | |
| variant_nucleotide_281 | |
| variant_protein_281 | |
| variant_gene_282 | |
| variant_nucleotide_282 | |
| variant_protein_282 | |
| variant_gene_283 | |
| variant_nucleotide_283 | |
| variant_protein_283 | |
| variant_gene_284 | |
| variant_nucleotide_284 | |
| variant_protein_284 | |
| variant_gene_285 | |
| variant_nucleotide_285 | |
| variant_protein_285 | |
| variant_gene_286 | |
| variant_nucleotide_286 | |
| variant_protein_286 | |
| variant_gene_287 | |
| variant_nucleotide_287 | |
| variant_protein_287 | |
| variant_gene_288 | |
| variant_nucleotide_288 | |
| variant_protein_288 | |
| variant_gene_289 | |
| variant_nucleotide_289 | |
| variant_protein_289 | |
| variant_gene_290 | |
| variant_nucleotide_290 | |
| variant_protein_290 | |
| variant_gene_291 | |
| variant_nucleotide_291 | |
| variant_protein_291 | |
| variant_gene_292 | |
| variant_nucleotide_292 | |
| variant_protein_292 | |
| variant_gene_293 | |
| variant_nucleotide_293 | |
| variant_protein_293 | |
| variant_gene_294 | |
| variant_nucleotide_294 | |
| variant_protein_294 | |
| variant_gene_295 | |
| variant_nucleotide_295 | |
| variant_protein_295 | |
| variant_gene_296 | |
| variant_nucleotide_296 | |
| variant_protein_296 | |
| variant_gene_297 | |
| variant_nucleotide_297 | |
| variant_protein_297 | |
| variant_gene_298 | |
| variant_nucleotide_298 | |
| variant_protein_298 | |
| variant_gene_299 | |
| variant_nucleotide_299 | |
| variant_protein_299 | |
| variant_gene_300 | |
| variant_nucleotide_300 | |
| variant_protein_300 | |
| variant_gene_301 | |
| variant_nucleotide_301 | |
| variant_protein_301 | |
| variant_gene_302 | |
| variant_nucleotide_302 | |
| variant_protein_302 | |
| variant_gene_303 | |
| variant_nucleotide_303 | |
| variant_protein_303 | |
| variant_gene_304 | |
| variant_nucleotide_304 | |
| variant_protein_304 | |
| variant_gene_305 | |
| variant_nucleotide_305 | |
| variant_protein_305 | |
| variant_gene_306 | |
| variant_nucleotide_306 | |
| variant_protein_306 | |
| variant_gene_307 | |
| variant_nucleotide_307 | |
| variant_protein_307 | |
| variant_gene_308 | |
| variant_nucleotide_308 | |
| variant_protein_308 | |
| variant_gene_309 | |
| variant_nucleotide_309 | |
| variant_protein_309 | |
| variant_gene_310 | |
| variant_nucleotide_310 | |
| variant_protein_310 | |
| variant_gene_311 | |
| variant_nucleotide_311 | |
| variant_protein_311 | |
| variant_gene_312 | |
| variant_nucleotide_312 | |
| variant_protein_312 | |
| variant_gene_313 | |
| variant_nucleotide_313 | |
| variant_protein_313 | |
| variant_gene_314 | |
| variant_nucleotide_314 | |
| variant_protein_314 | |
| variant_gene_315 | |
| variant_nucleotide_315 | |
| variant_protein_315 | |
| variant_gene_316 | |
| variant_nucleotide_316 | |
| variant_protein_316 | |
| variant_gene_317 | |
| variant_nucleotide_317 | |
| variant_protein_317 | |
| variant_gene_318 | |
| variant_nucleotide_318 | |
| variant_protein_318 | |
| variant_gene_319 | |
| variant_nucleotide_319 | |
| variant_protein_319 | |
| variant_gene_320 | |
| variant_nucleotide_320 | |
| variant_protein_320 | |
| variant_gene_321 | |
| variant_nucleotide_321 | |
| variant_protein_321 | |
| variant_gene_322 | |
| variant_nucleotide_322 | |
| variant_protein_322 | |
| variant_gene_323 | |
| variant_nucleotide_323 | |
| variant_protein_323 | |
| variant_gene_324 | |
| variant_nucleotide_324 | |
| variant_protein_324 | |
| variant_gene_325 | |
| variant_nucleotide_325 | |
| variant_protein_325 | |
| variant_gene_326 | |
| variant_nucleotide_326 | |
| variant_protein_326 | |
| variant_gene_327 | |
| variant_nucleotide_327 | |
| variant_protein_327 | |
| variant_gene_328 | |
| variant_nucleotide_328 | |
| variant_protein_328 | |
| variant_gene_329 | |
| variant_nucleotide_329 | |
| variant_protein_329 | |
| variant_gene_330 | |
| variant_nucleotide_330 | |
| variant_protein_330 | |
| variant_gene_331 | |
| variant_nucleotide_331 | |
| variant_protein_331 | |
| variant_gene_332 | |
| variant_nucleotide_332 | |
| variant_protein_332 | |
| variant_gene_333 | |
| variant_nucleotide_333 | |
| variant_protein_333 | |
| variant_gene_334 | |
| variant_nucleotide_334 | |
| variant_protein_334 | |
| variant_gene_335 | |
| variant_nucleotide_335 | |
| variant_protein_335 | |
| variant_gene_336 | |
| variant_nucleotide_336 | |
| variant_protein_336 | |
| variant_gene_337 | |
| variant_nucleotide_337 | |
| variant_protein_337 | |
| variant_gene_338 | |
| variant_nucleotide_338 | |
| variant_protein_338 | |
| variant_gene_339 | |
| variant_nucleotide_339 | |
| variant_protein_339 | |
| variant_gene_340 | |
| variant_nucleotide_340 | |
| variant_protein_340 | |
| variant_gene_341 | |
| variant_nucleotide_341 | |
| variant_protein_341 | |
| variant_gene_342 | |
| variant_nucleotide_342 | |
| variant_protein_342 | |
| variant_gene_343 | |
| variant_nucleotide_343 | |
| variant_protein_343 | |
| variant_gene_344 | |
| variant_nucleotide_344 | |
| variant_protein_344 | |
| variant_gene_345 | |
| variant_nucleotide_345 | |
| variant_protein_345 | |
| variant_gene_346 | |
| variant_nucleotide_346 | |
| variant_protein_346 | |
| variant_gene_347 | |
| variant_nucleotide_347 | |
| variant_protein_347 | |
| variant_gene_348 | |
| variant_nucleotide_348 | |
| variant_protein_348 | |
| variant_gene_349 | |
| variant_nucleotide_349 | |
| variant_protein_349 | |
| variant_gene_350 | |
| variant_nucleotide_350 | |
| variant_protein_350 | |
| variant_gene_351 | |
| variant_nucleotide_351 | |
| variant_protein_351 | |
| variant_gene_352 | |
| variant_nucleotide_352 | |
| variant_protein_352 | |
| variant_gene_353 | |
| variant_nucleotide_353 | |
| variant_protein_353 | |
| variant_gene_354 | |
| variant_nucleotide_354 | |
| variant_protein_354 | |
| variant_gene_355 | |
| variant_nucleotide_355 | |
| variant_protein_355 | |
| variant_gene_356 | |
| variant_nucleotide_356 | |
| variant_protein_356 | |
| variant_gene_357 | |
| variant_nucleotide_357 | |
| variant_protein_357 | |
| variant_gene_358 | |
| variant_nucleotide_358 | |
| variant_protein_358 | |
| variant_gene_359 | |
| variant_nucleotide_359 | |
| variant_protein_359 | |
| variant_gene_360 | |
| variant_nucleotide_360 | |
| variant_protein_360 | |
| variant_gene_361 | |
| variant_nucleotide_361 | |
| variant_protein_361 | |
| variant_gene_362 | |
| variant_nucleotide_362 | |
| variant_protein_362 | |
| variant_gene_363 | |
| variant_nucleotide_363 | |
| variant_protein_363 | |
| variant_gene_364 | |
| variant_nucleotide_364 | |
| variant_protein_364 | |
| variant_gene_365 | |
| variant_nucleotide_365 | |
| variant_protein_365 | |
| variant_gene_366 | |
| variant_nucleotide_366 | |
| variant_protein_366 | |
| variant_gene_367 | |
| variant_nucleotide_367 | |
| variant_protein_367 | |
| variant_gene_368 | |
| variant_nucleotide_368 | |
| variant_protein_368 | |
| variant_gene_369 | |
| variant_nucleotide_369 | |
| variant_protein_369 | |
| variant_gene_370 | |
| variant_nucleotide_370 | |
| variant_protein_370 | |
| variant_gene_371 | |
| variant_nucleotide_371 | |
| variant_protein_371 | |
| variant_gene_372 | |
| variant_nucleotide_372 | |
| variant_protein_372 | |
| variant_gene_373 | |
| variant_nucleotide_373 | |
| variant_protein_373 | |
| variant_gene_374 | |
| variant_nucleotide_374 | |
| variant_protein_374 | |
| variant_gene_375 | |
| variant_nucleotide_375 | |
| variant_protein_375 | |
| variant_gene_376 | |
| variant_nucleotide_376 | |
| variant_protein_376 | |
| variant_gene_377 | |
| variant_nucleotide_377 | |
| variant_protein_377 | |
| variant_gene_378 | |
| variant_nucleotide_378 | |
| variant_protein_378 | |
| variant_gene_379 | |
| variant_nucleotide_379 | |
| variant_protein_379 | |
| variant_gene_380 | |
| variant_nucleotide_380 | |
| variant_protein_380 | |
| variant_gene_381 | |
| variant_nucleotide_381 | |
| variant_protein_381 | |
| variant_gene_382 | |
| variant_nucleotide_382 | |
| variant_protein_382 | |
| variant_gene_383 | |
| variant_nucleotide_383 | |
| variant_protein_383 | |
| variant_gene_384 | |
| variant_nucleotide_384 | |
| variant_protein_384 | |
| variant_gene_385 | |
| variant_nucleotide_385 | |
| variant_protein_385 | |
| variant_gene_386 | |
| variant_nucleotide_386 | |
| variant_protein_386 | |
| variant_gene_387 | |
| variant_nucleotide_387 | |
| variant_protein_387 | |
| variant_gene_388 | |
| variant_nucleotide_388 | |
| variant_protein_388 | |
| variant_gene_389 | |
| variant_nucleotide_389 | |
| variant_protein_389 | |
| variant_gene_390 | |
| variant_nucleotide_390 | |
| variant_protein_390 | |
| variant_gene_391 | |
| variant_nucleotide_391 | |
| variant_protein_391 | |
| variant_gene_392 | |
| variant_nucleotide_392 | |
| variant_protein_392 | |
| variant_gene_393 | |
| variant_nucleotide_393 | |
| variant_protein_393 | |
| variant_gene_394 | |
| variant_nucleotide_394 | |
| variant_protein_394 | |
| variant_gene_395 | |
| variant_nucleotide_395 | |
| variant_protein_395 | |
| variant_gene_396 | |
| variant_nucleotide_396 | |
| variant_protein_396 | |
| variant_gene_397 | |
| variant_nucleotide_397 | |
| variant_protein_397 | |
| variant_gene_398 | |
| variant_nucleotide_398 | |
| variant_protein_398 | |
| variant_gene_399 | |
| variant_nucleotide_399 | |
| variant_protein_399 | |
| variant_gene_400 | |
| variant_nucleotide_400 | |
| variant_protein_400 | |
| variant_gene_401 | |
| variant_nucleotide_401 | |
| variant_protein_401 | |
| variant_gene_402 | |
| variant_nucleotide_402 | |
| variant_protein_402 | |
| variant_gene_403 | |
| variant_nucleotide_403 | |
| variant_protein_403 | |
| variant_gene_404 | |
| variant_nucleotide_404 | |
| variant_protein_404 | |
| variant_gene_405 | |
| variant_nucleotide_405 | |
| variant_protein_405 | |
| variant_gene_406 | |
| variant_nucleotide_406 | |
| variant_protein_406 | |
| variant_gene_407 | |
| variant_nucleotide_407 | |
| variant_protein_407 | |
| variant_gene_408 | |
| variant_nucleotide_408 | |
| variant_protein_408 | |
| variant_gene_409 | |
| variant_nucleotide_409 | |
| variant_protein_409 | |
| variant_gene_410 | |
| variant_nucleotide_410 | |
| variant_protein_410 | |
| variant_gene_411 | |
| variant_nucleotide_411 | |
| variant_protein_411 | |
| variant_gene_412 | |
| variant_nucleotide_412 | |
| variant_protein_412 | |
| variant_gene_413 | |
| variant_nucleotide_413 | |
| variant_protein_413 | |
| variant_gene_414 | |
| variant_nucleotide_414 | |
| variant_protein_414 | |
| variant_gene_415 | |
| variant_nucleotide_415 | |
| variant_protein_415 | |
| variant_gene_416 | |
| variant_nucleotide_416 | |
| variant_protein_416 | |
| variant_gene_417 | |
| variant_nucleotide_417 | |
| variant_protein_417 | |
| variant_gene_418 | |
| variant_nucleotide_418 | |
| variant_protein_418 | |
| variant_gene_419 | |
| variant_nucleotide_419 | |
| variant_protein_419 | |
| variant_gene_420 | |
| variant_nucleotide_420 | |
| variant_protein_420 | |
| variant_gene_421 | |
| variant_nucleotide_421 | |
| variant_protein_421 | |
| variant_gene_422 | |
| variant_nucleotide_422 | |
| variant_protein_422 | |
| variant_gene_423 | |
| variant_nucleotide_423 | |
| variant_protein_423 | |
| variant_gene_424 | |
| variant_nucleotide_424 | |
| variant_protein_424 | |
| variant_gene_425 | |
| variant_nucleotide_425 | |
| variant_protein_425 | |
| variant_gene_426 | |
| variant_nucleotide_426 | |
| variant_protein_426 | |
| variant_gene_427 | |
| variant_nucleotide_427 | |
| variant_protein_427 | |
| variant_gene_428 | |
| variant_nucleotide_428 | |
| variant_protein_428 | |
| variant_gene_429 | |
| variant_nucleotide_429 | |
| variant_protein_429 | |
| variant_gene_430 | |
| variant_nucleotide_430 | |
| variant_protein_430 | |
| variant_gene_431 | |
| variant_nucleotide_431 | |
| variant_protein_431 | |
| variant_gene_432 | |
| variant_nucleotide_432 | |
| variant_protein_432 | |
| variant_gene_433 | |
| variant_nucleotide_433 | |
| variant_protein_433 | |
| variant_gene_434 | |
| variant_nucleotide_434 | |
| variant_protein_434 | |
| variant_gene_435 | |
| variant_nucleotide_435 | |
| variant_protein_435 | |
| variant_gene_436 | |
| variant_nucleotide_436 | |
| variant_protein_436 | |
| variant_gene_437 | |
| variant_nucleotide_437 | |
| variant_protein_437 | |
| variant_gene_438 | |
| variant_nucleotide_438 | |
| variant_protein_438 | |
| variant_gene_439 | |
| variant_nucleotide_439 | |
| variant_protein_439 | |
| variant_gene_440 | |
| variant_nucleotide_440 | |
| variant_protein_440 | |
| variant_gene_441 | |
| variant_nucleotide_441 | |
| variant_protein_441 | |
| variant_gene_442 | |
| variant_nucleotide_442 | |
| variant_protein_442 | |
| variant_gene_443 | |
| variant_nucleotide_443 | |
| variant_protein_443 | |
| variant_gene_444 | |
| variant_nucleotide_444 | |
| variant_protein_444 | |
| variant_gene_445 | |
| variant_nucleotide_445 | |
| variant_protein_445 | |
| variant_gene_446 | |
| variant_nucleotide_446 | |
| variant_protein_446 | |
| variant_gene_447 | |
| variant_nucleotide_447 | |
| variant_protein_447 | |
| variant_gene_448 | |
| variant_nucleotide_448 | |
| variant_protein_448 | |
| variant_gene_449 | |
| variant_nucleotide_449 | |
| variant_protein_449 | |
| variant_gene_450 | |
| variant_nucleotide_450 | |
| variant_protein_450 | |
| variant_gene_451 | |
| variant_nucleotide_451 | |
| variant_protein_451 | |
| variant_gene_452 | |
| variant_nucleotide_452 | |
| variant_protein_452 | |
| variant_gene_453 | |
| variant_nucleotide_453 | |
| variant_protein_453 | |
| variant_gene_454 | |
| variant_nucleotide_454 | |
| variant_protein_454 | |
| variant_gene_455 | |
| variant_nucleotide_455 | |
| variant_protein_455 | |
| variant_gene_456 | |
| variant_nucleotide_456 | |
| variant_protein_456 | |
| variant_gene_457 | |
| variant_nucleotide_457 | |
| variant_protein_457 | |
| variant_gene_458 | |
| variant_nucleotide_458 | |
| variant_protein_458 | |
| variant_gene_459 | |
| variant_nucleotide_459 | |
| variant_protein_459 | |
| variant_gene_460 | |
| variant_nucleotide_460 | |
| variant_protein_460 | |
| variant_gene_461 | |
| variant_nucleotide_461 | |
| variant_protein_461 | |
| variant_gene_462 | |
| variant_nucleotide_462 | |
| variant_protein_462 | |
| variant_gene_463 | |
| variant_nucleotide_463 | |
| variant_protein_463 | |
| variant_gene_464 | |
| variant_nucleotide_464 | |
| variant_protein_464 | |
| variant_gene_465 | |
| variant_nucleotide_465 | |
| variant_protein_465 | |
| variant_gene_466 | |
| variant_nucleotide_466 | |
| variant_protein_466 | |
| variant_gene_467 | |
| variant_nucleotide_467 | |
| variant_protein_467 | |
| variant_gene_468 | |
| variant_nucleotide_468 | |
| variant_protein_468 | |
| variant_gene_469 | |
| variant_nucleotide_469 | |
| variant_protein_469 | |
| variant_gene_470 | |
| variant_nucleotide_470 | |
| variant_protein_470 | |
| variant_gene_471 | |
| variant_nucleotide_471 | |
| variant_protein_471 | |
| variant_gene_472 | |
| variant_nucleotide_472 | |
| variant_protein_472 | |
| variant_gene_473 | |
| variant_nucleotide_473 | |
| variant_protein_473 | |
| variant_gene_474 | |
| variant_nucleotide_474 | |
| variant_protein_474 | |
| variant_gene_475 | |
| variant_nucleotide_475 | |
| variant_protein_475 | |
| variant_gene_476 | |
| variant_nucleotide_476 | |
| variant_protein_476 | |
| variant_gene_477 | |
| variant_nucleotide_477 | |
| variant_protein_477 | |
| variant_gene_478 | |
| variant_nucleotide_478 | |
| variant_protein_478 | |
| variant_gene_479 | |
| variant_nucleotide_479 | |
| variant_protein_479 | |
| variant_gene_480 | |
| variant_nucleotide_480 | |
| variant_protein_480 | |
| variant_gene_481 | |
| variant_nucleotide_481 | |
| variant_protein_481 | |
| variant_gene_482 | |
| variant_nucleotide_482 | |
| variant_protein_482 | |
| variant_gene_483 | |
| variant_nucleotide_483 | |
| variant_protein_483 | |
| variant_gene_484 | |
| variant_nucleotide_484 | |
| variant_protein_484 | |
| variant_gene_485 | |
| variant_nucleotide_485 | |
| variant_protein_485 | |
| variant_gene_486 | |
| variant_nucleotide_486 | |
| variant_protein_486 | |
| variant_gene_487 | |
| variant_nucleotide_487 | |
| variant_protein_487 | |
| variant_gene_488 | |
| variant_nucleotide_488 | |
| variant_protein_488 | |
| variant_gene_489 | |
| variant_nucleotide_489 | |
| variant_protein_489 | |
| variant_gene_490 | |
| variant_nucleotide_490 | |
| variant_protein_490 | |
| variant_gene_491 | |
| variant_nucleotide_491 | |
| variant_protein_491 | |
| variant_gene_492 | |
| variant_nucleotide_492 | |
| variant_protein_492 | |
| variant_gene_493 | |
| variant_nucleotide_493 | |
| variant_protein_493 | |
| variant_gene_494 | |
| variant_nucleotide_494 | |
| variant_protein_494 | |
| variant_gene_495 | |
| variant_nucleotide_495 | |
| variant_protein_495 | |
| variant_gene_496 | |
| variant_nucleotide_496 | |
| variant_protein_496 | |
| variant_gene_497 | |
| variant_nucleotide_497 | |
| variant_protein_497 | |
| variant_gene_498 | |
| variant_nucleotide_498 | |
| variant_protein_498 | |
| variant_gene_499 | |
| variant_nucleotide_499 | |
| variant_protein_499 | |
| variant_gene_500 | |
| variant_nucleotide_500 | |
| variant_protein_500 | |
| variant_gene_501 | |
| variant_nucleotide_501 | |
| variant_protein_501 | |
| variant_gene_502 | |
| variant_nucleotide_502 | |
| variant_protein_502 | |
| variant_gene_503 | |
| variant_nucleotide_503 | |
| variant_protein_503 | |
| variant_gene_504 | |
| variant_nucleotide_504 | |
| variant_protein_504 | |
| variant_gene_505 | |
| variant_nucleotide_505 | |
| variant_protein_505 | |
| variant_gene_506 | |
| variant_nucleotide_506 | |
| variant_protein_506 | |
| variant_gene_507 | |
| variant_nucleotide_507 | |
| variant_protein_507 | |
| variant_gene_508 | |
| variant_nucleotide_508 | |
| variant_protein_508 | |
| variant_gene_509 | |
| variant_nucleotide_509 | |
| variant_protein_509 | |
| variant_gene_510 | |
| variant_nucleotide_510 | |
| variant_protein_510 | |
| variant_gene_511 | |
| variant_nucleotide_511 | |
| variant_protein_511 | |
| variant_gene_512 | |
| variant_nucleotide_512 | |
| variant_protein_512 | |
| variant_gene_513 | |
| variant_nucleotide_513 | |
| variant_protein_513 | |
| variant_gene_514 | |
| variant_nucleotide_514 | |
| variant_protein_514 | |
| variant_gene_515 | |
| variant_nucleotide_515 | |
| variant_protein_515 | |
| variant_gene_516 | |
| variant_nucleotide_516 | |
| variant_protein_516 | |
| variant_gene_517 | |
| variant_nucleotide_517 | |
| variant_protein_517 | |
| variant_gene_518 | |
| variant_nucleotide_518 | |
| variant_protein_518 | |
| variant_gene_519 | |
| variant_nucleotide_519 | |
| variant_protein_519 | |
| variant_gene_520 | |
| variant_nucleotide_520 | |
| variant_protein_520 | |
| variant_gene_521 | |
| variant_nucleotide_521 | |
| variant_protein_521 | |
| variant_gene_522 | |
| variant_nucleotide_522 | |
| variant_protein_522 | |
| variant_gene_523 | |
| variant_nucleotide_523 | |
| variant_protein_523 | |
| variant_gene_524 | |
| variant_nucleotide_524 | |
| variant_protein_524 | |
| variant_gene_525 | |
| variant_nucleotide_525 | |
| variant_protein_525 | |
| variant_gene_526 | |
| variant_nucleotide_526 | |
| variant_protein_526 | |
| variant_gene_527 | |
| variant_nucleotide_527 | |
| variant_protein_527 | |
| variant_gene_528 | |
| variant_nucleotide_528 | |
| variant_protein_528 | |
| variant_gene_529 | |
| variant_nucleotide_529 | |
| variant_protein_529 | |
| variant_gene_530 | |
| variant_nucleotide_530 | |
| variant_protein_530 | |
| variant_gene_531 | |
| variant_nucleotide_531 | |
| variant_protein_531 | |
| variant_gene_532 | |
| variant_nucleotide_532 | |
| variant_protein_532 | |
| variant_gene_533 | |
| variant_nucleotide_533 | |
| variant_protein_533 | |
| variant_gene_534 | |
| variant_nucleotide_534 | |
| variant_protein_534 | |
| variant_gene_535 | |
| variant_nucleotide_535 | |
| variant_protein_535 | |
| variant_gene_536 | |
| variant_nucleotide_536 | |
| variant_protein_536 | |
| variant_gene_537 | |
| variant_nucleotide_537 | |
| variant_protein_537 | |
| variant_gene_538 | |
| variant_nucleotide_538 | |
| variant_protein_538 | |
| variant_gene_539 | |
| variant_nucleotide_539 | |
| variant_protein_539 | |
| variant_gene_540 | |
| variant_nucleotide_540 | |
| variant_protein_540 | |
| variant_gene_541 | |
| variant_nucleotide_541 | |
| variant_protein_541 | |
| variant_gene_542 | |
| variant_nucleotide_542 | |
| variant_protein_542 | |
| variant_gene_543 | |
| variant_nucleotide_543 | |
| variant_protein_543 | |
| variant_gene_544 | |
| variant_nucleotide_544 | |
| variant_protein_544 | |
| variant_gene_545 | |
| variant_nucleotide_545 | |
| variant_protein_545 | |
| variant_gene_546 | |
| variant_nucleotide_546 | |
| variant_protein_546 | |
| variant_gene_547 | |
| variant_nucleotide_547 | |
| variant_protein_547 | |
| variant_gene_548 | |
| variant_nucleotide_548 | |
| variant_protein_548 | |
| variant_gene_549 | |
| variant_nucleotide_549 | |
| variant_protein_549 | |
| variant_gene_550 | |
| variant_nucleotide_550 | |
| variant_protein_550 | |
| variant_gene_551 | |
| variant_nucleotide_551 | |
| variant_protein_551 | |
| variant_gene_552 | |
| variant_nucleotide_552 | |
| variant_protein_552 | |
| variant_gene_553 | |
| variant_nucleotide_553 | |
| variant_protein_553 | |
| variant_gene_554 | |
| variant_nucleotide_554 | |
| variant_protein_554 | |
| variant_gene_555 | |
| variant_nucleotide_555 | |
| variant_protein_555 | |
| variant_gene_556 | |
| variant_nucleotide_556 | |
| variant_protein_556 | |
| variant_gene_557 | |
| variant_nucleotide_557 | |
| variant_protein_557 | |
| variant_gene_558 | |
| variant_nucleotide_558 | |
| variant_protein_558 | |
| variant_gene_559 | |
| variant_nucleotide_559 | |
| variant_protein_559 | |
| variant_gene_560 | |
| variant_nucleotide_560 | |
| variant_protein_560 | |
| variant_gene_561 | |
| variant_nucleotide_561 | |
| variant_protein_561 | |
| variant_gene_562 | |
| variant_nucleotide_562 | |
| variant_protein_562 | |
| variant_gene_563 | |
| variant_nucleotide_563 | |
| variant_protein_563 | |
| variant_gene_564 | |
| variant_nucleotide_564 | |
| variant_protein_564 | |
| variant_gene_565 | |
| variant_nucleotide_565 | |
| variant_protein_565 | |
| variant_gene_566 | |
| variant_nucleotide_566 | |
| variant_protein_566 | |
| variant_gene_567 | |
| variant_nucleotide_567 | |
| variant_protein_567 | |
| variant_gene_568 | |
| variant_nucleotide_568 | |
| variant_protein_568 | |
| variant_gene_569 | |
| variant_nucleotide_569 | |
| variant_protein_569 | |
| variant_gene_570 | |
| variant_nucleotide_570 | |
| variant_protein_570 | |
| variant_gene_571 | |
| variant_nucleotide_571 | |
| variant_protein_571 | |
| variant_gene_572 | |
| variant_nucleotide_572 | |
| variant_protein_572 | |
| variant_gene_573 | |
| variant_nucleotide_573 | |
| variant_protein_573 | |
| variant_gene_574 | |
| variant_nucleotide_574 | |
| variant_protein_574 | |
| variant_gene_575 | |
| variant_nucleotide_575 | |
| variant_protein_575 | |
| variant_gene_576 | |
| variant_nucleotide_576 | |
| variant_protein_576 | |
| variant_gene_577 | |
| variant_nucleotide_577 | |
| variant_protein_577 | |
| variant_gene_578 | |
| variant_nucleotide_578 | |
| variant_protein_578 | |
| variant_gene_579 | |
| variant_nucleotide_579 | |
| variant_protein_579 | |
| variant_gene_580 | |
| variant_nucleotide_580 | |
| variant_protein_580 | |
| variant_gene_581 | |
| variant_nucleotide_581 | |
| variant_protein_581 | |
| variant_gene_582 | |
| variant_nucleotide_582 | |
| variant_protein_582 | |
| variant_gene_583 | |
| variant_nucleotide_583 | |
| variant_protein_583 | |
| variant_gene_584 | |
| variant_nucleotide_584 | |
| variant_protein_584 | |
| variant_gene_585 | |
| variant_nucleotide_585 | |
| variant_protein_585 | |
| variant_gene_586 | |
| variant_nucleotide_586 | |
| variant_protein_586 | |
| variant_gene_587 | |
| variant_nucleotide_587 | |
| variant_protein_587 | |
| variant_gene_588 | |
| variant_nucleotide_588 | |
| variant_protein_588 | |
| variant_gene_589 | |
| variant_nucleotide_589 | |
| variant_protein_589 | |
| variant_gene_590 | |
| variant_nucleotide_590 | |
| variant_protein_590 | |
| variant_gene_591 | |
| variant_nucleotide_591 | |
| variant_protein_591 | |
| variant_gene_592 | |
| variant_nucleotide_592 | |
| variant_protein_592 | |
| variant_gene_593 | |
| variant_nucleotide_593 | |
| variant_protein_593 | |
| variant_gene_594 | |
| variant_nucleotide_594 | |
| variant_protein_594 | |
| variant_gene_595 | |
| variant_nucleotide_595 | |
| variant_protein_595 | |
| variant_gene_596 | |
| variant_nucleotide_596 | |
| variant_protein_596 | |
| variant_gene_597 | |
| variant_nucleotide_597 | |
| variant_protein_597 | |
| variant_gene_598 | |
| variant_nucleotide_598 | |
| variant_protein_598 | |
| variant_gene_599 | |
| variant_nucleotide_599 | |
| variant_protein_599 | |
| variant_gene_600 | |
| variant_nucleotide_600 | |
| variant_protein_600 | |
| variant_gene_601 | |
| variant_nucleotide_601 | |
| variant_protein_601 | |
| variant_gene_602 | |
| variant_nucleotide_602 | |
| variant_protein_602 | |
| variant_gene_603 | |
| variant_nucleotide_603 | |
| variant_protein_603 | |
| variant_gene_604 | |
| variant_nucleotide_604 | |
| variant_protein_604 | |
| variant_gene_605 | |
| variant_nucleotide_605 | |
| variant_protein_605 | |
| variant_gene_606 | |
| variant_nucleotide_606 | |
| variant_protein_606 | |
| variant_gene_607 | |
| variant_nucleotide_607 | |
| variant_protein_607 | |
| variant_gene_608 | |
| variant_nucleotide_608 | |
| variant_protein_608 | |
| variant_gene_609 | |
| variant_nucleotide_609 | |
| variant_protein_609 | |
| variant_gene_610 | |
| variant_nucleotide_610 | |
| variant_protein_610 | |
| variant_gene_611 | |
| variant_nucleotide_611 | |
| variant_protein_611 | |
| variant_gene_612 | |
| variant_nucleotide_612 | |
| variant_protein_612 | |
| variant_gene_613 | |
| variant_nucleotide_613 | |
| variant_protein_613 | |
| variant_gene_614 | |
| variant_nucleotide_614 | |
| variant_protein_614 | |
| variant_gene_615 | |
| variant_nucleotide_615 | |
| variant_protein_615 | |
| variant_gene_616 | |
| variant_nucleotide_616 | |
| variant_protein_616 | |
| variant_gene_617 | |
| variant_nucleotide_617 | |
| variant_protein_617 | |
| variant_gene_618 | |
| variant_nucleotide_618 | |
| variant_protein_618 | |
| variant_gene_619 | |
| variant_nucleotide_619 | |
| variant_protein_619 | |
| variant_gene_620 | |
| variant_nucleotide_620 | |
| variant_protein_620 | |
| variant_gene_621 | |
| variant_nucleotide_621 | |
| variant_protein_621 | |
| variant_gene_622 | |
| variant_nucleotide_622 | |
| variant_protein_622 | |
| variant_gene_623 | |
| variant_nucleotide_623 | |
| variant_protein_623 | |
| variant_gene_624 | |
| variant_nucleotide_624 | |
| variant_protein_624 | |
| variant_gene_625 | |
| variant_nucleotide_625 | |
| variant_protein_625 | |
| variant_gene_626 | |
| variant_nucleotide_626 | |
| variant_protein_626 | |
| variant_gene_627 | |
| variant_nucleotide_627 | |
| variant_protein_627 | |
| variant_gene_628 | |
| variant_nucleotide_628 | |
| variant_protein_628 | |
| variant_gene_629 | |
| variant_nucleotide_629 | |
| variant_protein_629 | |
| variant_gene_630 | |
| variant_nucleotide_630 | |
| variant_protein_630 | |
| variant_gene_631 | |
| variant_nucleotide_631 | |
| variant_protein_631 | |
| variant_gene_632 | |
| variant_nucleotide_632 | |
| variant_protein_632 | |
| variant_gene_633 | |
| variant_nucleotide_633 | |
| variant_protein_633 | |
| variant_gene_634 | |
| variant_nucleotide_634 | |
| variant_protein_634 | |
| variant_gene_635 | |
| variant_nucleotide_635 | |
| variant_protein_635 | |
| variant_gene_636 | |
| variant_nucleotide_636 | |
| variant_protein_636 | |
| variant_gene_637 | |
| variant_nucleotide_637 | |
| variant_protein_637 | |
| variant_gene_638 | |
| variant_nucleotide_638 | |
| variant_protein_638 | |
| variant_gene_639 | |
| variant_nucleotide_639 | |
| variant_protein_639 | |
| variant_gene_640 | |
| variant_nucleotide_640 | |
| variant_protein_640 | |
| variant_gene_641 | |
| variant_nucleotide_641 | |
| variant_protein_641 | |
| variant_gene_642 | |
| variant_nucleotide_642 | |
| variant_protein_642 | |
| variant_gene_643 | |
| variant_nucleotide_643 | |
| variant_protein_643 | |
| variant_gene_644 | |
| variant_nucleotide_644 | |
| variant_protein_644 | |
| variant_gene_645 | |
| variant_nucleotide_645 | |
| variant_protein_645 | |
| variant_gene_646 | |
| variant_nucleotide_646 | |
| variant_protein_646 | |
| variant_gene_647 | |
| variant_nucleotide_647 | |
| variant_protein_647 | |
| variant_gene_648 | |
| variant_nucleotide_648 | |
| variant_protein_648 | |
| variant_gene_649 | |
| variant_nucleotide_649 | |
| variant_protein_649 | |
| variant_gene_650 | |
| variant_nucleotide_650 | |
| variant_protein_650 | |
| variant_gene_651 | |
| variant_nucleotide_651 | |
| variant_protein_651 | |
| variant_gene_652 | |
| variant_nucleotide_652 | |
| variant_protein_652 | |
| variant_gene_653 | |
| variant_nucleotide_653 | |
| variant_protein_653 | |
| variant_gene_654 | |
| variant_nucleotide_654 | |
| variant_protein_654 | |
| variant_gene_655 | |
| variant_nucleotide_655 | |
| variant_protein_655 | |
| variant_gene_656 | |
| variant_nucleotide_656 | |
| variant_protein_656 | |
| variant_gene_657 | |
| variant_nucleotide_657 | |
| variant_protein_657 | |
| variant_gene_658 | |
| variant_nucleotide_658 | |
| variant_protein_658 | |
| variant_gene_659 | |
| variant_nucleotide_659 | |
| variant_protein_659 | |
| variant_gene_660 | |
| variant_nucleotide_660 | |
| variant_protein_660 | |
| variant_gene_661 | |
| variant_nucleotide_661 | |
| variant_protein_661 | |
| variant_gene_662 | |
| variant_nucleotide_662 | |
| variant_protein_662 | |
| variant_gene_663 | |
| variant_nucleotide_663 | |
| variant_protein_663 | |
| variant_gene_664 | |
| variant_nucleotide_664 | |
| variant_protein_664 | |
| variant_gene_665 | |
| variant_nucleotide_665 | |
| variant_protein_665 | |
| variant_gene_666 | |
| variant_nucleotide_666 | |
| variant_protein_666 | |
| variant_gene_667 | |
| variant_nucleotide_667 | |
| variant_protein_667 | |
| variant_gene_668 | |
| variant_nucleotide_668 | |
| variant_protein_668 | |
| variant_gene_669 | |
| variant_nucleotide_669 | |
| variant_protein_669 | |
| variant_gene_670 | |
| variant_nucleotide_670 | |
| variant_protein_670 | |
| variant_gene_671 | |
| variant_nucleotide_671 | |
| variant_protein_671 | |
| variant_gene_672 | |
| variant_nucleotide_672 | |
| variant_protein_672 | |
| variant_gene_673 | |
| variant_nucleotide_673 | |
| variant_protein_673 | |
| variant_gene_674 | |
| variant_nucleotide_674 | |
| variant_protein_674 | |
| variant_gene_675 | |
| variant_nucleotide_675 | |
| variant_protein_675 | |
| variant_gene_676 | |
| variant_nucleotide_676 | |
| variant_protein_676 | |
| variant_gene_677 | |
| variant_nucleotide_677 | |
| variant_protein_677 | |
| variant_gene_678 | |
| variant_nucleotide_678 | |
| variant_protein_678 | |
| variant_gene_679 | |
| variant_nucleotide_679 | |
| variant_protein_679 | |
| variant_gene_680 | |
| variant_nucleotide_680 | |
| variant_protein_680 | |
| variant_gene_681 | |
| variant_nucleotide_681 | |
| variant_protein_681 | |
| variant_gene_682 | |
| variant_nucleotide_682 | |
| variant_protein_682 | |
| variant_gene_683 | |
| variant_nucleotide_683 | |
| variant_protein_683 | |
| variant_gene_684 | |
| variant_nucleotide_684 | |
| variant_protein_684 | |
| variant_gene_685 | |
| variant_nucleotide_685 | |
| variant_protein_685 | |
| variant_gene_686 | |
| variant_nucleotide_686 | |
| variant_protein_686 | |
| variant_gene_687 | |
| variant_nucleotide_687 | |
| variant_protein_687 | |
| variant_gene_688 | |
| variant_nucleotide_688 | |
| variant_protein_688 | |
| variant_gene_689 | |
| variant_nucleotide_689 | |
| variant_protein_689 | |
| variant_gene_690 | |
| variant_nucleotide_690 | |
| variant_protein_690 | |
| variant_gene_691 | |
| variant_nucleotide_691 | |
| variant_protein_691 | |
| variant_gene_692 | |
| variant_nucleotide_692 | |
| variant_protein_692 | |
| variant_gene_693 | |
| variant_nucleotide_693 | |
| variant_protein_693 | |
| variant_gene_694 | |
| variant_nucleotide_694 | |
| variant_protein_694 | |
| variant_gene_695 | |
| variant_nucleotide_695 | |
| variant_protein_695 | |
| variant_gene_696 | |
| variant_nucleotide_696 | |
| variant_protein_696 | |
| variant_gene_697 | |
| variant_nucleotide_697 | |
| variant_protein_697 | |
| variant_gene_698 | |
| variant_nucleotide_698 | |
| variant_protein_698 | |
| variant_gene_699 | |
| variant_nucleotide_699 | |
| variant_protein_699 | |
| variant_gene_700 | |
| variant_nucleotide_700 | |
| variant_protein_700 | |
| variant_gene_701 | |
| variant_nucleotide_701 | |
| variant_protein_701 | |
| variant_gene_702 | |
| variant_nucleotide_702 | |
| variant_protein_702 | |
| variant_gene_703 | |
| variant_nucleotide_703 | |
| variant_protein_703 | |
| variant_gene_704 | |
| variant_nucleotide_704 | |
| variant_protein_704 | |
| variant_gene_705 | |
| variant_nucleotide_705 | |
| variant_protein_705 | |
| variant_gene_706 | |
| variant_nucleotide_706 | |
| variant_protein_706 | |
| variant_gene_707 | |
| variant_nucleotide_707 | |
| variant_protein_707 | |
| variant_gene_708 | |
| variant_nucleotide_708 | |
| variant_protein_708 | |
| variant_gene_709 | |
| variant_nucleotide_709 | |
| variant_protein_709 | |
| variant_gene_710 | |
| variant_nucleotide_710 | |
| variant_protein_710 | |
| variant_gene_711 | |
| variant_nucleotide_711 | |
| variant_protein_711 | |
| variant_gene_712 | |
| variant_nucleotide_712 | |
| variant_protein_712 | |
| variant_gene_713 | |
| variant_nucleotide_713 | |
| variant_protein_713 | |
| variant_gene_714 | |
| variant_nucleotide_714 | |
| variant_protein_714 | |
| variant_gene_715 | |
| variant_nucleotide_715 | |
| variant_protein_715 | |
| variant_gene_716 | |
| variant_nucleotide_716 | |
| variant_protein_716 | |
| variant_gene_717 | |
| variant_nucleotide_717 | |
| variant_protein_717 | |
| variant_gene_718 | |
| variant_nucleotide_718 | |
| variant_protein_718 | |
| variant_gene_719 | |
| variant_nucleotide_719 | |
| variant_protein_719 | |
| variant_gene_720 | |
| variant_nucleotide_720 | |
| variant_protein_720 | |
| variant_gene_721 | |
| variant_nucleotide_721 | |
| variant_protein_721 | |
| variant_gene_722 | |
| variant_nucleotide_722 | |
| variant_protein_722 | |
| variant_gene_723 | |
| variant_nucleotide_723 | |
| variant_protein_723 | |
| variant_gene_724 | |
| variant_nucleotide_724 | |
| variant_protein_724 | |
| variant_gene_725 | |
| variant_nucleotide_725 | |
| variant_protein_725 | |
| variant_gene_726 | |
| variant_nucleotide_726 | |
| variant_protein_726 | |
| variant_gene_727 | |
| variant_nucleotide_727 | |
| variant_protein_727 | |
| variant_gene_728 | |
| variant_nucleotide_728 | |
| variant_protein_728 | |
| variant_gene_729 | |
| variant_nucleotide_729 | |
| variant_protein_729 | |
| variant_gene_730 | |
| variant_nucleotide_730 | |
| variant_protein_730 | |
| variant_gene_731 | |
| variant_nucleotide_731 | |
| variant_protein_731 | |
| variant_gene_732 | |
| variant_nucleotide_732 | |
| variant_protein_732 | |
| variant_gene_733 | |
| variant_nucleotide_733 | |
| variant_protein_733 | |
| variant_gene_734 | |
| variant_nucleotide_734 | |
| variant_protein_734 | |
| variant_gene_735 | |
| variant_nucleotide_735 | |
| variant_protein_735 | |
| variant_gene_736 | |
| variant_nucleotide_736 | |
| variant_protein_736 | |
| variant_gene_737 | |
| variant_nucleotide_737 | |
| variant_protein_737 | |
| variant_gene_738 | |
| variant_nucleotide_738 | |
| variant_protein_738 | |
| variant_gene_739 | |
| variant_nucleotide_739 | |
| variant_protein_739 | |
| variant_gene_740 | |
| variant_nucleotide_740 | |
| variant_protein_740 | |
| variant_gene_741 | |
| variant_nucleotide_741 | |
| variant_protein_741 | |
| variant_gene_742 | |
| variant_nucleotide_742 | |
| variant_protein_742 | |
| variant_gene_743 | |
| variant_nucleotide_743 | |
| variant_protein_743 | |
| variant_gene_744 | |
| variant_nucleotide_744 | |
| variant_protein_744 | |
| variant_gene_745 | |
| variant_nucleotide_745 | |
| variant_protein_745 | |
| variant_gene_746 | |
| variant_nucleotide_746 | |
| variant_protein_746 | |
| variant_gene_747 | |
| variant_nucleotide_747 | |
| variant_protein_747 | |
| variant_gene_748 | |
| variant_nucleotide_748 | |
| variant_protein_748 | |
| variant_gene_749 | |
| variant_nucleotide_749 | |
| variant_protein_749 | |
| variant_gene_750 | |
| variant_nucleotide_750 | |
| variant_protein_750 | |
| variant_gene_751 | |
| variant_nucleotide_751 | |
| variant_protein_751 | |
| variant_gene_752 | |
| variant_nucleotide_752 | |
| variant_protein_752 | |
| variant_gene_753 | |
| variant_nucleotide_753 | |
| variant_protein_753 | |
| variant_gene_754 | |
| variant_nucleotide_754 | |
| variant_protein_754 | |
| variant_gene_755 | |
| variant_nucleotide_755 | |
| variant_protein_755 | |
| variant_gene_756 | |
| variant_nucleotide_756 | |
| variant_protein_756 | |
| variant_gene_757 | |
| variant_nucleotide_757 | |
| variant_protein_757 | |
| variant_gene_758 | |
| variant_nucleotide_758 | |
| variant_protein_758 | |
| variant_gene_759 | |
| variant_nucleotide_759 | |
| variant_protein_759 | |
| variant_gene_760 | |
| variant_nucleotide_760 | |
| variant_protein_760 | |
| variant_gene_761 | |
| variant_nucleotide_761 | |
| variant_protein_761 | |
| variant_gene_762 | |
| variant_nucleotide_762 | |
| variant_protein_762 | |
| variant_gene_763 | |
| variant_nucleotide_763 | |
| variant_protein_763 | |
| variant_gene_764 | |
| variant_nucleotide_764 | |
| variant_protein_764 | |
| variant_gene_765 | |
| variant_nucleotide_765 | |
| variant_protein_765 | |
| variant_gene_766 | |
| variant_nucleotide_766 | |
| variant_protein_766 | |
| variant_gene_767 | |
| variant_nucleotide_767 | |
| variant_protein_767 | |
| variant_gene_768 | |
| variant_nucleotide_768 | |
| variant_protein_768 | |
| variant_gene_769 | |
| variant_nucleotide_769 | |
| variant_protein_769 | |
| variant_gene_770 | |
| variant_nucleotide_770 | |
| variant_protein_770 | |
| variant_gene_771 | |
| variant_nucleotide_771 | |
| variant_protein_771 | |
| variant_gene_772 | |
| variant_nucleotide_772 | |
| variant_protein_772 | |
| variant_gene_773 | |
| variant_nucleotide_773 | |
| variant_protein_773 | |
| variant_gene_774 | |
| variant_nucleotide_774 | |
| variant_protein_774 | |
| variant_gene_775 | |
| variant_nucleotide_775 | |
| variant_protein_775 | |
| variant_gene_776 | |
| variant_nucleotide_776 | |
| variant_protein_776 | |
| variant_gene_777 | |
| variant_nucleotide_777 | |
| variant_protein_777 | |
| variant_gene_778 | |
| variant_nucleotide_778 | |
| variant_protein_778 | |
| variant_gene_779 | |
| variant_nucleotide_779 | |
| variant_protein_779 | |
| variant_gene_780 | |
| variant_nucleotide_780 | |
| variant_protein_780 | |
| variant_gene_781 | |
| variant_nucleotide_781 | |
| variant_protein_781 | |
| variant_gene_782 | |
| variant_nucleotide_782 | |
| variant_protein_782 | |
| variant_gene_783 | |
| variant_nucleotide_783 | |
| variant_protein_783 | |
| variant_gene_784 | |
| variant_nucleotide_784 | |
| variant_protein_784 | |
| variant_gene_785 | |
| variant_nucleotide_785 | |
| variant_protein_785 | |
| variant_gene_786 | |
| variant_nucleotide_786 | |
| variant_protein_786 | |
| variant_gene_787 | |
| variant_nucleotide_787 | |
| variant_protein_787 | |
| variant_gene_788 | |
| variant_nucleotide_788 | |
| variant_protein_788 | |
| variant_gene_789 | |
| variant_nucleotide_789 | |
| variant_protein_789 | |
| variant_gene_790 | |
| variant_nucleotide_790 | |
| variant_protein_790 | |
| variant_gene_791 | |
| variant_nucleotide_791 | |
| variant_protein_791 | |
| variant_gene_792 | |
| variant_nucleotide_792 | |
| variant_protein_792 | |
| variant_gene_793 | |
| variant_nucleotide_793 | |
| variant_protein_793 | |
| variant_gene_794 | |
| variant_nucleotide_794 | |
| variant_protein_794 | |
| variant_gene_795 | |
| variant_nucleotide_795 | |
| variant_protein_795 | |
| variant_gene_796 | |
| variant_nucleotide_796 | |
| variant_protein_796 | |
| variant_gene_797 | |
| variant_nucleotide_797 | |
| variant_protein_797 | |
| variant_gene_798 | |
| variant_nucleotide_798 | |
| variant_protein_798 | |
| variant_gene_799 | |
| variant_nucleotide_799 | |
| variant_protein_799 | |
| variant_gene_800 | |
| variant_nucleotide_800 | |
| variant_protein_800 | |
| variant_gene_801 | |
| variant_nucleotide_801 | |
| variant_protein_801 | |
| variant_gene_802 | |
| variant_nucleotide_802 | |
| variant_protein_802 | |
| variant_gene_803 | |
| variant_nucleotide_803 | |
| variant_protein_803 | |
| variant_gene_804 | |
| variant_nucleotide_804 | |
| variant_protein_804 | |
| variant_gene_805 | |
| variant_nucleotide_805 | |
| variant_protein_805 | |
| variant_gene_806 | |
| variant_nucleotide_806 | |
| variant_protein_806 | |
| variant_gene_807 | |
| variant_nucleotide_807 | |
| variant_protein_807 | |
| variant_gene_808 | |
| variant_nucleotide_808 | |
| variant_protein_808 | |
| variant_gene_809 | |
| variant_nucleotide_809 | |
| variant_protein_809 | |
| variant_gene_810 | |
| variant_nucleotide_810 | |
| variant_protein_810 | |
| variant_gene_811 | |
| variant_nucleotide_811 | |
| variant_protein_811 | |
| variant_gene_812 | |
| variant_nucleotide_812 | |
| variant_protein_812 | |
| variant_gene_813 | |
| variant_nucleotide_813 | |
| variant_protein_813 | |
| variant_gene_814 | |
| variant_nucleotide_814 | |
| variant_protein_814 | |
| variant_gene_815 | |
| variant_nucleotide_815 | |
| variant_protein_815 | |
| variant_gene_816 | |
| variant_nucleotide_816 | |
| variant_protein_816 | |
| variant_gene_817 | |
| variant_nucleotide_817 | |
| variant_protein_817 | |
| variant_gene_818 | |
| variant_nucleotide_818 | |
| variant_protein_818 | |
| variant_gene_819 | |
| variant_nucleotide_819 | |
| variant_protein_819 | |
| variant_gene_820 | |
| variant_nucleotide_820 | |
| variant_protein_820 | |
| variant_gene_821 | |
| variant_nucleotide_821 | |
| variant_protein_821 | |
| variant_gene_822 | |
| variant_nucleotide_822 | |
| variant_protein_822 | |
| variant_gene_823 | |
| variant_nucleotide_823 | |
| variant_protein_823 | |
| variant_gene_824 | |
| variant_nucleotide_824 | |
| variant_protein_824 | |
| variant_gene_825 | |
| variant_nucleotide_825 | |
| variant_protein_825 | |
| variant_gene_826 | |
| variant_nucleotide_826 | |
| variant_protein_826 | |
| variant_gene_827 | |
| variant_nucleotide_827 | |
| variant_protein_827 | |
| variant_gene_828 | |
| variant_nucleotide_828 | |
| variant_protein_828 | |
| variant_gene_829 | |
| variant_nucleotide_829 | |
| variant_protein_829 | |
| variant_gene_830 | |
| variant_nucleotide_830 | |
| variant_protein_830 | |
| variant_gene_831 | |
| variant_nucleotide_831 | |
| variant_protein_831 | |
| variant_gene_832 | |
| variant_nucleotide_832 | |
| variant_protein_832 | |
| variant_gene_833 | |
| variant_nucleotide_833 | |
| variant_protein_833 | |
| variant_gene_834 | |
| variant_nucleotide_834 | |
| variant_protein_834 | |
| variant_gene_835 | |
| variant_nucleotide_835 | |
| variant_protein_835 | |
| variant_gene_836 | |
| variant_nucleotide_836 | |
| variant_protein_836 | |
| variant_gene_837 | |
| variant_nucleotide_837 | |
| variant_protein_837 | |
| variant_gene_838 | |
| variant_nucleotide_838 | |
| variant_protein_838 | |
| variant_gene_839 | |
| variant_nucleotide_839 | |
| variant_protein_839 | |
| variant_gene_840 | |
| variant_nucleotide_840 | |
| variant_protein_840 | |
| variant_gene_841 | |
| variant_nucleotide_841 | |
| variant_protein_841 | |
| variant_gene_842 | |
| variant_nucleotide_842 | |
| variant_protein_842 | |
| variant_gene_843 | |
| variant_nucleotide_843 | |
| variant_protein_843 | |
| variant_gene_844 | |
| variant_nucleotide_844 | |
| variant_protein_844 | |
| variant_gene_845 | |
| variant_nucleotide_845 | |
| variant_protein_845 | |
| variant_gene_846 | |
| variant_nucleotide_846 | |
| variant_protein_846 | |
| variant_gene_847 | |
| variant_nucleotide_847 | |
| variant_protein_847 | |
| variant_gene_848 | |
| variant_nucleotide_848 | |
| variant_protein_848 | |
| variant_gene_849 | |
| variant_nucleotide_849 | |
| variant_protein_849 | |
| variant_gene_850 | |
| variant_nucleotide_850 | |
| variant_protein_850 | |
| variant_gene_851 | |
| variant_nucleotide_851 | |
| variant_protein_851 | |
| variant_gene_852 | |
| variant_nucleotide_852 | |
| variant_protein_852 | |
| variant_gene_853 | |
| variant_nucleotide_853 | |
| variant_protein_853 | |
| variant_gene_854 | |
| variant_nucleotide_854 | |
| variant_protein_854 | |
| variant_gene_855 | |
| variant_nucleotide_855 | |
| variant_protein_855 | |
| variant_gene_856 | |
| variant_nucleotide_856 | |
| variant_protein_856 | |
| variant_gene_857 | |
| variant_nucleotide_857 | |
| variant_protein_857 | |
| variant_gene_858 | |
| variant_nucleotide_858 | |
| variant_protein_858 | |
| variant_gene_859 | |
| variant_nucleotide_859 | |
| variant_protein_859 | |
| variant_gene_860 | |
| variant_nucleotide_860 | |
| variant_protein_860 | |
| variant_gene_861 | |
| variant_nucleotide_861 | |
| variant_protein_861 | |
| variant_gene_862 | |
| variant_nucleotide_862 | |
| variant_protein_862 | |
| variant_gene_863 | |
| variant_nucleotide_863 | |
| variant_protein_863 | |
| variant_gene_864 | |
| variant_nucleotide_864 | |
| variant_protein_864 | |
| variant_gene_865 | |
| variant_nucleotide_865 | |
| variant_protein_865 | |
| variant_gene_866 | |
| variant_nucleotide_866 | |
| variant_protein_866 | |
| variant_gene_867 | |
| variant_nucleotide_867 | |
| variant_protein_867 | |
| variant_gene_868 | |
| variant_nucleotide_868 | |
| variant_protein_868 | |
| variant_gene_869 | |
| variant_nucleotide_869 | |
| variant_protein_869 | |
| variant_gene_870 | |
| variant_nucleotide_870 | |
| variant_protein_870 | |
| variant_gene_871 | |
| variant_nucleotide_871 | |
| variant_protein_871 | |
| variant_gene_872 | |
| variant_nucleotide_872 | |
| variant_protein_872 | |
| variant_gene_873 | |
| variant_nucleotide_873 | |
| variant_protein_873 | |
| variant_gene_874 | |
| variant_nucleotide_874 | |
| variant_protein_874 | |
| variant_gene_875 | |
| variant_nucleotide_875 | |
| variant_protein_875 | |
| variant_gene_876 | |
| variant_nucleotide_876 | |
| variant_protein_876 | |
| variant_gene_877 | |
| variant_nucleotide_877 | |
| variant_protein_877 | |
| variant_gene_878 | |
| variant_nucleotide_878 | |
| variant_protein_878 | |
| variant_gene_879 | |
| variant_nucleotide_879 | |
| variant_protein_879 | |
| variant_gene_880 | |
| variant_nucleotide_880 | |
| variant_protein_880 | |
| variant_gene_881 | |
| variant_nucleotide_881 | |
| variant_protein_881 | |
| variant_gene_882 | |
| variant_nucleotide_882 | |
| variant_protein_882 | |
| variant_gene_883 | |
| variant_nucleotide_883 | |
| variant_protein_883 | |
| variant_gene_884 | |
| variant_nucleotide_884 | |
| variant_protein_884 | |
| variant_gene_885 | |
| variant_nucleotide_885 | |
| variant_protein_885 | |
| variant_gene_886 | |
| variant_nucleotide_886 | |
| variant_protein_886 | |
| variant_gene_887 | |
| variant_nucleotide_887 | |
| variant_protein_887 | |
| variant_gene_888 | |
| variant_nucleotide_888 | |
| variant_protein_888 | |
| variant_gene_889 | |
| variant_nucleotide_889 | |
| variant_protein_889 | |
| variant_gene_890 | |
| variant_nucleotide_890 | |
| variant_protein_890 | |
| variant_gene_891 | |
| variant_nucleotide_891 | |
| variant_protein_891 | |
| variant_gene_892 | |
| variant_nucleotide_892 | |
| variant_protein_892 | |
| variant_gene_893 | |
| variant_nucleotide_893 | |
| variant_protein_893 | |
| variant_gene_894 | |
| variant_nucleotide_894 | |
| variant_protein_894 | |
| variant_gene_895 | |
| variant_nucleotide_895 | |
| variant_protein_895 | |
| variant_gene_896 | |
| variant_nucleotide_896 | |
| variant_protein_896 | |
| variant_gene_897 | |
| variant_nucleotide_897 | |
| variant_protein_897 | |
| variant_gene_898 | |
| variant_nucleotide_898 | |
| variant_protein_898 | |
| variant_gene_899 | |
| variant_nucleotide_899 | |
| variant_protein_899 | |
| variant_gene_900 | |
| variant_nucleotide_900 | |
| variant_protein_900 | |
| variant_gene_901 | |
| variant_nucleotide_901 | |
| variant_protein_901 | |
| variant_gene_902 | |
| variant_nucleotide_902 | |
| variant_protein_902 | |
| variant_gene_903 | |
| variant_nucleotide_903 | |
| variant_protein_903 | |
| variant_gene_904 | |
| variant_nucleotide_904 | |
| variant_protein_904 | |
| variant_gene_905 | |
| variant_nucleotide_905 | |
| variant_protein_905 | |
| variant_gene_906 | |
| variant_nucleotide_906 | |
| variant_protein_906 | |
| variant_gene_907 | |
| variant_nucleotide_907 | |
| variant_protein_907 | |
| variant_gene_908 | |
| variant_nucleotide_908 | |
| variant_protein_908 | |
| variant_gene_909 | |
| variant_nucleotide_909 | |
| variant_protein_909 | |
| variant_gene_910 | |
| variant_nucleotide_910 | |
| variant_protein_910 | |
| variant_gene_911 | |
| variant_nucleotide_911 | |
| variant_protein_911 | |
| variant_gene_912 | |
| variant_nucleotide_912 | |
| variant_protein_912 | |
| variant_gene_913 | |
| variant_nucleotide_913 | |
| variant_protein_913 | |
| variant_gene_914 | |
| variant_nucleotide_914 | |
| variant_protein_914 | |
| variant_gene_915 | |
| variant_nucleotide_915 | |
| variant_protein_915 | |
| variant_gene_916 | |
| variant_nucleotide_916 | |
| variant_protein_916 | |
| variant_gene_917 | |
| variant_nucleotide_917 | |
| variant_protein_917 | |
| variant_gene_918 | |
| variant_nucleotide_918 | |
| variant_protein_918 | |
| variant_gene_919 | |
| variant_nucleotide_919 | |
| variant_protein_919 | |
| variant_gene_920 | |
| variant_nucleotide_920 | |
| variant_protein_920 | |
| variant_gene_921 | |
| variant_nucleotide_921 | |
| variant_protein_921 | |
| variant_gene_922 | |
| variant_nucleotide_922 | |
| variant_protein_922 | |
| variant_gene_923 | |
| variant_nucleotide_923 | |
| variant_protein_923 | |
| variant_gene_924 | |
| variant_nucleotide_924 | |
| variant_protein_924 | |
| variant_gene_925 | |
| variant_nucleotide_925 | |
| variant_protein_925 | |
| variant_gene_926 | |
| variant_nucleotide_926 | |
| variant_protein_926 | |
| variant_gene_927 | |
| variant_nucleotide_927 | |
| variant_protein_927 | |
| variant_gene_928 | |
| variant_nucleotide_928 | |
| variant_protein_928 | |
| variant_gene_929 | |
| variant_nucleotide_929 | |
| variant_protein_929 | |
| variant_gene_930 | |
| variant_nucleotide_930 | |
| variant_protein_930 | |
| variant_gene_931 | |
| variant_nucleotide_931 | |
| variant_protein_931 | |
| variant_gene_932 | |
| variant_nucleotide_932 | |
| variant_protein_932 | |
| variant_gene_933 | |
| variant_nucleotide_933 | |
| variant_protein_933 | |
| variant_gene_934 | |
| variant_nucleotide_934 | |
| variant_protein_934 | |
| variant_gene_935 | |
| variant_nucleotide_935 | |
| variant_protein_935 | |
| variant_gene_936 | |
| variant_nucleotide_936 | |
| variant_protein_936 | |
| variant_gene_937 | |
| variant_nucleotide_937 | |
| variant_protein_937 | |
| variant_gene_938 | |
| variant_nucleotide_938 | |
| variant_protein_938 | |
| variant_gene_939 | |
| variant_nucleotide_939 | |
| variant_protein_939 | |
| variant_gene_940 | |
| variant_nucleotide_940 | |
| variant_protein_940 | |
| variant_gene_941 | |
| variant_nucleotide_941 | |
| variant_protein_941 | |
| variant_gene_942 | |
| variant_nucleotide_942 | |
| variant_protein_942 | |
| variant_gene_943 | |
| variant_nucleotide_943 | |
| variant_protein_943 | |
| variant_gene_944 | |
| variant_nucleotide_944 | |
| variant_protein_944 | |
| variant_gene_945 | |
| variant_nucleotide_945 | |
| variant_protein_945 | |
| variant_gene_946 | |
| variant_nucleotide_946 | |
| variant_protein_946 | |
| variant_gene_947 | |
| variant_nucleotide_947 | |
| variant_protein_947 | |
| variant_gene_948 | |
| variant_nucleotide_948 | |
| variant_protein_948 | |
| variant_gene_949 | |
| variant_nucleotide_949 | |
| variant_protein_949 | |
| variant_gene_950 | |
| variant_nucleotide_950 | |
| variant_protein_950 | |
| variant_gene_951 | |
| variant_nucleotide_951 | |
| variant_protein_951 | |
| variant_gene_952 | |
| variant_nucleotide_952 | |
| variant_protein_952 | |
| variant_gene_953 | |
| variant_nucleotide_953 | |
| variant_protein_953 | |
| variant_gene_954 | |
| variant_nucleotide_954 | |
| variant_protein_954 | |
| variant_gene_955 | |
| variant_nucleotide_955 | |
| variant_protein_955 | |
| variant_gene_956 | |
| variant_nucleotide_956 | |
| variant_protein_956 | |
| variant_gene_957 | |
| variant_nucleotide_957 | |
| variant_protein_957 | |
| variant_gene_958 | |
| variant_nucleotide_958 | |
| variant_protein_958 | |
| variant_gene_959 | |
| variant_nucleotide_959 | |
| variant_protein_959 | |
| variant_gene_960 | |
| variant_nucleotide_960 | |
| variant_protein_960 | |
| variant_gene_961 | |
| variant_nucleotide_961 | |
| variant_protein_961 | |
| variant_gene_962 | |
| variant_nucleotide_962 | |
| variant_protein_962 | |
| variant_gene_963 | |
| variant_nucleotide_963 | |
| variant_protein_963 | |
| variant_gene_964 | |
| variant_nucleotide_964 | |
| variant_protein_964 | |
| variant_gene_965 | |
| variant_nucleotide_965 | |
| variant_protein_965 | |
| variant_gene_966 | |
| variant_nucleotide_966 | |
| variant_protein_966 | |
| variant_gene_967 | |
| variant_nucleotide_967 | |
| variant_protein_967 | |
| variant_gene_968 | |
| variant_nucleotide_968 | |
| variant_protein_968 | |
| variant_gene_969 | |
| variant_nucleotide_969 | |
| variant_protein_969 | |
| variant_gene_970 | |
| variant_nucleotide_970 | |
| variant_protein_970 | |
| variant_gene_971 | |
| variant_nucleotide_971 | |
| variant_protein_971 | |
| variant_gene_972 | |
| variant_nucleotide_972 | |
| variant_protein_972 | |
| variant_gene_973 | |
| variant_nucleotide_973 | |
| variant_protein_973 | |
| variant_gene_974 | |
| variant_nucleotide_974 | |
| variant_protein_974 | |
| variant_gene_975 | |
| variant_nucleotide_975 | |
| variant_protein_975 | |
| variant_gene_976 | |
| variant_nucleotide_976 | |
| variant_protein_976 | |
| variant_gene_977 | |
| variant_nucleotide_977 | |
| variant_protein_977 | |
| variant_gene_978 | |
| variant_nucleotide_978 | |
| variant_protein_978 | |
| variant_gene_979 | |
| variant_nucleotide_979 | |
| variant_protein_979 | |
| variant_gene_980 | |
| variant_nucleotide_980 | |
| variant_protein_980 | |
| variant_gene_981 | |
| variant_nucleotide_981 | |
| variant_protein_981 | |
| variant_gene_982 | |
| variant_nucleotide_982 | |
| variant_protein_982 | |
| variant_gene_983 | |
| variant_nucleotide_983 | |
| variant_protein_983 | |
| variant_gene_984 | |
| variant_nucleotide_984 | |
| variant_protein_984 | |
| variant_gene_985 | |
| variant_nucleotide_985 | |
| variant_protein_985 | |
| variant_gene_986 | |
| variant_nucleotide_986 | |
| variant_protein_986 | |
| variant_gene_987 | |
| variant_nucleotide_987 | |
| variant_protein_987 | |
| variant_gene_988 | |
| variant_nucleotide_988 | |
| variant_protein_988 | |
| variant_gene_989 | |
| variant_nucleotide_989 | |
| variant_protein_989 | |
| variant_gene_990 | |
| variant_nucleotide_990 | |
| variant_protein_990 | |
| variant_gene_991 | |
| variant_nucleotide_991 | |
| variant_protein_991 | |
| variant_gene_992 | |
| variant_nucleotide_992 | |
| variant_protein_992 | |
| variant_gene_993 | |
| variant_nucleotide_993 | |
| variant_protein_993 | |
| variant_gene_994 | |
| variant_nucleotide_994 | |
| variant_protein_994 | |
| variant_gene_995 | |
| variant_nucleotide_995 | |
| variant_protein_995 | |
| variant_gene_996 | |
| variant_nucleotide_996 | |
| variant_protein_996 | |
| variant_gene_997 | |
| variant_nucleotide_997 | |
| variant_protein_997 | |
| variant_gene_998 | |
| variant_nucleotide_998 | |
| variant_protein_998 | |
| variant_gene_999 | |
| variant_nucleotide_999 | |
| variant_protein_999 | |
| variant_gene_1000 | |
| variant_nucleotide_1000 | |
| variant_protein_1000 | |
| variant_gene_1001 | |
| variant_nucleotide_1001 | |
| variant_protein_1001 | |
| variant_gene_1002 | |
| variant_nucleotide_1002 | |
| variant_protein_1002 | |
| variant_gene_1003 | |
| variant_nucleotide_1003 | |
| variant_protein_1003 | |
| variant_gene_1004 | |
| variant_nucleotide_1004 | |
| variant_protein_1004 | |
| variant_gene_1005 | |
| variant_nucleotide_1005 | |
| variant_protein_1005 | |
| variant_gene_1006 | |
| variant_nucleotide_1006 | |
| variant_protein_1006 | |
| variant_gene_1007 | |
| variant_nucleotide_1007 | |
| variant_protein_1007 | |
| variant_gene_1008 | |
| variant_nucleotide_1008 | |
| variant_protein_1008 | |
| variant_gene_1009 | |
| variant_nucleotide_1009 | |
| variant_protein_1009 | |
| variant_gene_1010 | |
| variant_nucleotide_1010 | |
| variant_protein_1010 | |
| variant_gene_1011 | |
| variant_nucleotide_1011 | |
| variant_protein_1011 | |
| variant_gene_1012 | |
| variant_nucleotide_1012 | |
| variant_protein_1012 | |
| variant_gene_1013 | |
| variant_nucleotide_1013 | |
| variant_protein_1013 | |
| variant_gene_1014 | |
| variant_nucleotide_1014 | |
| variant_protein_1014 | |
| variant_gene_1015 | |
| variant_nucleotide_1015 | |
| variant_protein_1015 | |
| variant_gene_1016 | |
| variant_nucleotide_1016 | |
| variant_protein_1016 | |
| variant_gene_1017 | |
| variant_nucleotide_1017 | |
| variant_protein_1017 | |
| variant_gene_1018 | |
| variant_nucleotide_1018 | |
| variant_protein_1018 | |
| variant_gene_1019 | |
| variant_nucleotide_1019 | |
| variant_protein_1019 | |
| variant_gene_1020 | |
| variant_nucleotide_1020 | |
| variant_protein_1020 | |
| variant_gene_1021 | |
| variant_nucleotide_1021 | |
| variant_protein_1021 | |
| variant_gene_1022 | |
| variant_nucleotide_1022 | |
| variant_protein_1022 | |
| variant_gene_1023 | |
| variant_nucleotide_1023 | |
| variant_protein_1023 | |
| variant_gene_1024 | |
| variant_nucleotide_1024 | |
| variant_protein_1024 | |
| variant_gene_1025 | |
| variant_nucleotide_1025 | |
| variant_protein_1025 | |
| variant_gene_1026 | |
| variant_nucleotide_1026 | |
| variant_protein_1026 | |
| variant_gene_1027 | |
| variant_nucleotide_1027 | |
| variant_protein_1027 | |
| variant_gene_1028 | |
| variant_nucleotide_1028 | |
| variant_protein_1028 | |
| variant_gene_1029 | |
| variant_nucleotide_1029 | |
| variant_protein_1029 | |
| variant_gene_1030 | |
| variant_nucleotide_1030 | |
| variant_protein_1030 | |
| variant_gene_1031 | |
| variant_nucleotide_1031 | |
| variant_protein_1031 | |
| variant_gene_1032 | |
| variant_nucleotide_1032 | |
| variant_protein_1032 | |
| variant_gene_1033 | |
| variant_nucleotide_1033 | |
| variant_protein_1033 | |
| variant_gene_1034 | |
| variant_nucleotide_1034 | |
| variant_protein_1034 | |
| variant_gene_1035 | |
| variant_nucleotide_1035 | |
| variant_protein_1035 | |
| variant_gene_1036 | |
| variant_nucleotide_1036 | |
| variant_protein_1036 | |
| variant_gene_1037 | |
| variant_nucleotide_1037 | |
| variant_protein_1037 | |
| variant_gene_1038 | |
| variant_nucleotide_1038 | |
| variant_protein_1038 | |
| variant_gene_1039 | |
| variant_nucleotide_1039 | |
| variant_protein_1039 | |
| variant_gene_1040 | |
| variant_nucleotide_1040 | |
| variant_protein_1040 | |
| variant_gene_1041 | |
| variant_nucleotide_1041 | |
| variant_protein_1041 | |
| variant_gene_1042 | |
| variant_nucleotide_1042 | |
| variant_protein_1042 | |
| variant_gene_1043 | |
| variant_nucleotide_1043 | |
| variant_protein_1043 | |
| variant_gene_1044 | |
| variant_nucleotide_1044 | |
| variant_protein_1044 | |
| variant_gene_1045 | |
| variant_nucleotide_1045 | |
| variant_protein_1045 | |
| variant_gene_1046 | |
| variant_nucleotide_1046 | |
| variant_protein_1046 | |
| variant_gene_1047 | |
| variant_nucleotide_1047 | |
| variant_protein_1047 | |
| variant_gene_1048 | |
| variant_nucleotide_1048 | |
| variant_protein_1048 | |
| variant_gene_1049 | |
| variant_nucleotide_1049 | |
| variant_protein_1049 | |
| variant_gene_1050 | |
| variant_nucleotide_1050 | |
| variant_protein_1050 | |
| variant_gene_1051 | |
| variant_nucleotide_1051 | |
| variant_protein_1051 | |
| variant_gene_1052 | |
| variant_nucleotide_1052 | |
| variant_protein_1052 | |
| variant_gene_1053 | |
| variant_nucleotide_1053 | |
| variant_protein_1053 | |
| variant_gene_1054 | |
| variant_nucleotide_1054 | |
| variant_protein_1054 | |
| variant_gene_1055 | |
| variant_nucleotide_1055 | |
| variant_protein_1055 | |
| variant_gene_1056 | |
| variant_nucleotide_1056 | |
| variant_protein_1056 | |
| variant_gene_1057 | |
| variant_nucleotide_1057 | |
| variant_protein_1057 | |
| variant_gene_1058 | |
| variant_nucleotide_1058 | |
| variant_protein_1058 | |
| variant_gene_1059 | |
| variant_nucleotide_1059 | |
| variant_protein_1059 | |
| variant_gene_1060 | |
| variant_nucleotide_1060 | |
| variant_protein_1060 | |
| variant_gene_1061 | |
| variant_nucleotide_1061 | |
| variant_protein_1061 | |
| variant_gene_1062 | |
| variant_nucleotide_1062 | |
| variant_protein_1062 | |
| variant_gene_1063 | |
| variant_nucleotide_1063 | |
| variant_protein_1063 | |
| variant_gene_1064 | |
| variant_nucleotide_1064 | |
| variant_protein_1064 | |
| variant_gene_1065 | |
| variant_nucleotide_1065 | |
| variant_protein_1065 | |
| variant_gene_1066 | |
| variant_nucleotide_1066 | |
| variant_protein_1066 | |
| variant_gene_1067 | |
| variant_nucleotide_1067 | |
| variant_protein_1067 | |
| variant_gene_1068 | |
| variant_nucleotide_1068 | |
| variant_protein_1068 | |
| variant_gene_1069 | |
| variant_nucleotide_1069 | |
| variant_protein_1069 | |
| variant_gene_1070 | |
| variant_nucleotide_1070 | |
| variant_protein_1070 | |
| variant_gene_1071 | |
| variant_nucleotide_1071 | |
| variant_protein_1071 | |
| variant_gene_1072 | |
| variant_nucleotide_1072 | |
| variant_protein_1072 | |
| variant_gene_1073 | |
| variant_nucleotide_1073 | |
| variant_protein_1073 | |
| variant_gene_1074 | |
| variant_nucleotide_1074 | |
| variant_protein_1074 | |
| variant_gene_1075 | |
| variant_nucleotide_1075 | |
| variant_protein_1075 | |
| variant_gene_1076 | |
| variant_nucleotide_1076 | |
| variant_protein_1076 | |
| variant_gene_1077 | |
| variant_nucleotide_1077 | |
| variant_protein_1077 | |
| variant_gene_1078 | |
| variant_nucleotide_1078 | |
| variant_protein_1078 | |
| variant_gene_1079 | |
| variant_nucleotide_1079 | |
| variant_protein_1079 | |
| variant_gene_1080 | |
| variant_nucleotide_1080 | |
| variant_protein_1080 | |
| variant_gene_1081 | |
| variant_nucleotide_1081 | |
| variant_protein_1081 | |
| variant_gene_1082 | |
| variant_nucleotide_1082 | |
| variant_protein_1082 | |
| variant_gene_1083 | |
| variant_nucleotide_1083 | |
| variant_protein_1083 | |
| variant_gene_1084 | |
| variant_nucleotide_1084 | |
| variant_protein_1084 | |
| variant_gene_1085 | |
| variant_nucleotide_1085 | |
| variant_protein_1085 | |
| variant_gene_1086 | |
| variant_nucleotide_1086 | |
| variant_protein_1086 | |
| variant_gene_1087 | |
| variant_nucleotide_1087 | |
| variant_protein_1087 | |
| variant_gene_1088 | |
| variant_nucleotide_1088 | |
| variant_protein_1088 | |
| variant_gene_1089 | |
| variant_nucleotide_1089 | |
| variant_protein_1089 | |
| variant_gene_1090 | |
| variant_nucleotide_1090 | |
| variant_protein_1090 | |
| variant_gene_1091 | |
| variant_nucleotide_1091 | |
| variant_protein_1091 | |
| variant_gene_1092 | |
| variant_nucleotide_1092 | |
| variant_protein_1092 | |
| variant_gene_1093 | |
| variant_nucleotide_1093 | |
| variant_protein_1093 | |
| variant_gene_1094 | |
| variant_nucleotide_1094 | |
| variant_protein_1094 | |
| variant_gene_1095 | |
| variant_nucleotide_1095 | |
| variant_protein_1095 | |
| variant_gene_1096 | |
| variant_nucleotide_1096 | |
| variant_protein_1096 | |
| variant_gene_1097 | |
| variant_nucleotide_1097 | |
| variant_protein_1097 | |
| variant_gene_1098 | |
| variant_nucleotide_1098 | |
| variant_protein_1098 | |
| variant_gene_1099 | |
| variant_nucleotide_1099 | |
| variant_protein_1099 | |
| variant_gene_1100 | |
| variant_nucleotide_1100 | |
| variant_protein_1100 | |
| variant_gene_1101 | |
| variant_nucleotide_1101 | |
| variant_protein_1101 | |
| variant_gene_1102 | |
| variant_nucleotide_1102 | |
| variant_protein_1102 | |
| variant_gene_1103 | |
| variant_nucleotide_1103 | |
| variant_protein_1103 | |
| variant_gene_1104 | |
| variant_nucleotide_1104 | |
| variant_protein_1104 | |
| variant_gene_1105 | |
| variant_nucleotide_1105 | |
| variant_protein_1105 | |
| variant_gene_1106 | |
| variant_nucleotide_1106 | |
| variant_protein_1106 | |
| variant_gene_1107 | |
| variant_nucleotide_1107 | |
| variant_protein_1107 | |
| variant_gene_1108 | |
| variant_nucleotide_1108 | |
| variant_protein_1108 | |
| variant_gene_1109 | |
| variant_nucleotide_1109 | |
| variant_protein_1109 | |
| variant_gene_1110 | |
| variant_nucleotide_1110 | |
| variant_protein_1110 | |
| variant_gene_1111 | |
| variant_nucleotide_1111 | |
| variant_protein_1111 | |
| variant_gene_1112 | |
| variant_nucleotide_1112 | |
| variant_protein_1112 | |
| variant_gene_1113 | |
| variant_nucleotide_1113 | |
| variant_protein_1113 | |
| variant_gene_1114 | |
| variant_nucleotide_1114 | |
| variant_protein_1114 | |
| variant_gene_1115 | |
| variant_nucleotide_1115 | |
| variant_protein_1115 | |
| variant_gene_1116 | |
| variant_nucleotide_1116 | |
| variant_protein_1116 | |
| variant_gene_1117 | |
| variant_nucleotide_1117 | |
| variant_protein_1117 | |
| variant_gene_1118 | |
| variant_nucleotide_1118 | |
| variant_protein_1118 | |
| variant_gene_1119 | |
| variant_nucleotide_1119 | |
| variant_protein_1119 | |
| variant_gene_1120 | |
| variant_nucleotide_1120 | |
| variant_protein_1120 | |
| variant_gene_1121 | |
| variant_nucleotide_1121 | |
| variant_protein_1121 | |
| variant_gene_1122 | |
| variant_nucleotide_1122 | |
| variant_protein_1122 | |
| variant_gene_1123 | |
| variant_nucleotide_1123 | |
| variant_protein_1123 | |
| variant_gene_1124 | |
| variant_nucleotide_1124 | |
| variant_protein_1124 | |
| variant_gene_1125 | |
| variant_nucleotide_1125 | |
| variant_protein_1125 | |
| variant_gene_1126 | |
| variant_nucleotide_1126 | |
| variant_protein_1126 | |
| variant_gene_1127 | |
| variant_nucleotide_1127 | |
| variant_protein_1127 | |
| variant_gene_1128 | |
| variant_nucleotide_1128 | |
| variant_protein_1128 | |
| variant_gene_1129 | |
| variant_nucleotide_1129 | |
| variant_protein_1129 | |
| variant_gene_1130 | |
| variant_nucleotide_1130 | |
| variant_protein_1130 | |
| variant_gene_1131 | |
| variant_nucleotide_1131 | |
| variant_protein_1131 | |
| variant_gene_1132 | |
| variant_nucleotide_1132 | |
| variant_protein_1132 | |
| variant_gene_1133 | |
| variant_nucleotide_1133 | |
| variant_protein_1133 | |
| variant_gene_1134 | |
| variant_nucleotide_1134 | |
| variant_protein_1134 | |
| variant_gene_1135 | |
| variant_nucleotide_1135 | |
| variant_protein_1135 | |
| variant_gene_1136 | |
| variant_nucleotide_1136 | |
| variant_protein_1136 | |
| variant_gene_1137 | |
| variant_nucleotide_1137 | |
| variant_protein_1137 | |
| variant_gene_1138 | |
| variant_nucleotide_1138 | |
| variant_protein_1138 | |
| variant_gene_1139 | |
| variant_nucleotide_1139 | |
| variant_protein_1139 | |
| variant_gene_1140 | |
| variant_nucleotide_1140 | |
| variant_protein_1140 | |
| variant_gene_1141 | |
| variant_nucleotide_1141 | |
| variant_protein_1141 | |
| variant_gene_1142 | |
| variant_nucleotide_1142 | |
| variant_protein_1142 | |
| variant_gene_1143 | |
| variant_nucleotide_1143 | |
| variant_protein_1143 | |
| variant_gene_1144 | |
| variant_nucleotide_1144 | |
| variant_protein_1144 | |
| variant_gene_1145 | |
| variant_nucleotide_1145 | |
| variant_protein_1145 | |
| variant_gene_1146 | |
| variant_nucleotide_1146 | |
| variant_protein_1146 | |
| variant_gene_1147 | |
| variant_nucleotide_1147 | |
| variant_protein_1147 | |
| variant_gene_1148 | |
| variant_nucleotide_1148 | |
| variant_protein_1148 | |
| variant_gene_1149 | |
| variant_nucleotide_1149 | |
| variant_protein_1149 | |
| variant_gene_1150 | |
| variant_nucleotide_1150 | |
| variant_protein_1150 | |
| variant_gene_1151 | |
| variant_nucleotide_1151 | |
| variant_protein_1151 | |
| variant_gene_1152 | |
| variant_nucleotide_1152 | |
| variant_protein_1152 | |
| variant_gene_1153 | |
| variant_nucleotide_1153 | |
| variant_protein_1153 | |
| variant_gene_1154 | |
| variant_nucleotide_1154 | |
| variant_protein_1154 | |
| variant_gene_1155 | |
| variant_nucleotide_1155 | |
| variant_protein_1155 | |
| variant_gene_1156 | |
| variant_nucleotide_1156 | |
| variant_protein_1156 | |
| variant_gene_1157 | |
| variant_nucleotide_1157 | |
| variant_protein_1157 | |
| variant_gene_1158 | |
| variant_nucleotide_1158 | |
| variant_protein_1158 | |
| variant_gene_1159 | |
| variant_nucleotide_1159 | |
| variant_protein_1159 | |
| variant_gene_1160 | |
| variant_nucleotide_1160 | |
| variant_protein_1160 | |
| variant_gene_1161 | |
| variant_nucleotide_1161 | |
| variant_protein_1161 | |
| variant_gene_1162 | |
| variant_nucleotide_1162 | |
| variant_protein_1162 | |
| variant_gene_1163 | |
| variant_nucleotide_1163 | |
| variant_protein_1163 | |
| variant_gene_1164 | |
| variant_nucleotide_1164 | |
| variant_protein_1164 | |
| variant_gene_1165 | |
| variant_nucleotide_1165 | |
| variant_protein_1165 | |
| variant_gene_1166 | |
| variant_nucleotide_1166 | |
| variant_protein_1166 | |
| variant_gene_1167 | |
| variant_nucleotide_1167 | |
| variant_protein_1167 | |
| variant_gene_1168 | |
| variant_nucleotide_1168 | |
| variant_protein_1168 | |
| variant_gene_1169 | |
| variant_nucleotide_1169 | |
| variant_protein_1169 | |
| variant_gene_1170 | |
| variant_nucleotide_1170 | |
| variant_protein_1170 | |
| variant_gene_1171 | |
| variant_nucleotide_1171 | |
| variant_protein_1171 | |
| variant_gene_1172 | |
| variant_nucleotide_1172 | |
| variant_protein_1172 | |
| variant_gene_1173 | |
| variant_nucleotide_1173 | |
| variant_protein_1173 | |
| variant_gene_1174 | |
| variant_nucleotide_1174 | |
| variant_protein_1174 | |
| variant_gene_1175 | |
| variant_nucleotide_1175 | |
| variant_protein_1175 | |
| variant_gene_1176 | |
| variant_nucleotide_1176 | |
| variant_protein_1176 | |
| variant_gene_1177 | |
| variant_nucleotide_1177 | |
| variant_protein_1177 | |
| variant_gene_1178 | |
| variant_nucleotide_1178 | |
| variant_protein_1178 | |
| variant_gene_1179 | |
| variant_nucleotide_1179 | |
| variant_protein_1179 | |
| variant_gene_1180 | |
| variant_nucleotide_1180 | |
| variant_protein_1180 | |
| variant_gene_1181 | |
| variant_nucleotide_1181 | |
| variant_protein_1181 | |
| variant_gene_1182 | |
| variant_nucleotide_1182 | |
| variant_protein_1182 | |
| variant_gene_1183 | |
| variant_nucleotide_1183 | |
| variant_protein_1183 | |
| variant_gene_1184 | |
| variant_nucleotide_1184 | |
| variant_protein_1184 | |
| variant_gene_1185 | |
| variant_nucleotide_1185 | |
| variant_protein_1185 | |
| variant_gene_1186 | |
| variant_nucleotide_1186 | |
| variant_protein_1186 | |
| variant_gene_1187 | |
| variant_nucleotide_1187 | |
| variant_protein_1187 | |
| variant_gene_1188 | |
| variant_nucleotide_1188 | |
| variant_protein_1188 | |
| variant_gene_1189 | |
| variant_nucleotide_1189 | |
| variant_protein_1189 | |
| variant_gene_1190 | |
| variant_nucleotide_1190 | |
| variant_protein_1190 | |
| variant_gene_1191 | |
| variant_nucleotide_1191 | |
| variant_protein_1191 | |
| variant_gene_1192 | |
| variant_nucleotide_1192 | |
| variant_protein_1192 | |
| variant_gene_1193 | |
| variant_nucleotide_1193 | |
| variant_protein_1193 | |
| variant_gene_1194 | |
| variant_nucleotide_1194 | |
| variant_protein_1194 | |
| variant_gene_1195 | |
| variant_nucleotide_1195 | |
| variant_protein_1195 | |
| variant_gene_1196 | |
| variant_nucleotide_1196 | |
| variant_protein_1196 | |
| variant_gene_1197 | |
| variant_nucleotide_1197 | |
| variant_protein_1197 | |
| variant_gene_1198 | |
| variant_nucleotide_1198 | |
| variant_protein_1198 | |
| variant_gene_1199 | |
| variant_nucleotide_1199 | |
| variant_protein_1199 | |
| variant_gene_1200 | |
| variant_nucleotide_1200 | |
| variant_protein_1200 | |
| variant_gene_1201 | |
| variant_nucleotide_1201 | |
| variant_protein_1201 | |
| variant_gene_1202 | |
| variant_nucleotide_1202 | |
| variant_protein_1202 | |
| variant_gene_1203 | |
| variant_nucleotide_1203 | |
| variant_protein_1203 | |
| variant_gene_1204 | |
| variant_nucleotide_1204 | |
| variant_protein_1204 | |
| variant_gene_1205 | |
| variant_nucleotide_1205 | |
| variant_protein_1205 | |
| variant_gene_1206 | |
| variant_nucleotide_1206 | |
| variant_protein_1206 | |
| variant_gene_1207 | |
| variant_nucleotide_1207 | |
| variant_protein_1207 | |
| variant_gene_1208 | |
| variant_nucleotide_1208 | |
| variant_protein_1208 | |
| variant_gene_1209 | |
| variant_nucleotide_1209 | |
| variant_protein_1209 | |
| variant_gene_1210 | |
| variant_nucleotide_1210 | |
| variant_protein_1210 | |
| variant_gene_1211 | |
| variant_nucleotide_1211 | |
| variant_protein_1211 | |
| variant_gene_1212 | |
| variant_nucleotide_1212 | |
| variant_protein_1212 | |
| variant_gene_1213 | |
| variant_nucleotide_1213 | |
| variant_protein_1213 | |
| variant_gene_1214 | |
| variant_nucleotide_1214 | |
| variant_protein_1214 | |
| variant_gene_1215 | |
| variant_nucleotide_1215 | |
| variant_protein_1215 | |
| variant_gene_1216 | |
| variant_nucleotide_1216 | |
| variant_protein_1216 | |
| variant_gene_1217 | |
| variant_nucleotide_1217 | |
| variant_protein_1217 | |
| variant_gene_1218 | |
| variant_nucleotide_1218 | |
| variant_protein_1218 | |
| variant_gene_1219 | |
| variant_nucleotide_1219 | |
| variant_protein_1219 | |
| variant_gene_1220 | |
| variant_nucleotide_1220 | |
| variant_protein_1220 | |
| variant_gene_1221 | |
| variant_nucleotide_1221 | |
| variant_protein_1221 | |
| variant_gene_1222 | |
| variant_nucleotide_1222 | |
| variant_protein_1222 | |
| variant_gene_1223 | |
| variant_nucleotide_1223 | |
| variant_protein_1223 | |
| variant_gene_1224 | |
| variant_nucleotide_1224 | |
| variant_protein_1224 | |
| variant_gene_1225 | |
| variant_nucleotide_1225 | |
| variant_protein_1225 | |
| variant_gene_1226 | |
| variant_nucleotide_1226 | |
| variant_protein_1226 | |
| variant_gene_1227 | |
| variant_nucleotide_1227 | |
| variant_protein_1227 | |
| variant_gene_1228 | |
| variant_nucleotide_1228 | |
| variant_protein_1228 | |
| variant_gene_1229 | |
| variant_nucleotide_1229 | |
| variant_protein_1229 | |
| variant_gene_1230 | |
| variant_nucleotide_1230 | |
| variant_protein_1230 | |
| variant_gene_1231 | |
| variant_nucleotide_1231 | |
| variant_protein_1231 | |
| variant_gene_1232 | |
| variant_nucleotide_1232 | |
| variant_protein_1232 | |
| variant_gene_1233 | |
| variant_nucleotide_1233 | |
| variant_protein_1233 | |
| variant_gene_1234 | |
| variant_nucleotide_1234 | |
| variant_protein_1234 | |
| variant_gene_1235 | |
| variant_nucleotide_1235 | |
| variant_protein_1235 | |
| variant_gene_1236 | |
| variant_nucleotide_1236 | |
| variant_protein_1236 | |
| variant_gene_1237 | |
| variant_nucleotide_1237 | |
| variant_protein_1237 | |
| variant_gene_1238 | |
| variant_nucleotide_1238 | |
| variant_protein_1238 | |
| variant_gene_1239 | |
| variant_nucleotide_1239 | |
| variant_protein_1239 | |
| variant_gene_1240 | |
| variant_nucleotide_1240 | |
| variant_protein_1240 | |
| variant_gene_1241 | |
| variant_nucleotide_1241 | |
| variant_protein_1241 | |
| variant_gene_1242 | |
| variant_nucleotide_1242 | |
| variant_protein_1242 | |
| variant_gene_1243 | |
| variant_nucleotide_1243 | |
| variant_protein_1243 | |
| variant_gene_1244 | |
| variant_nucleotide_1244 | |
| variant_protein_1244 | |
| variant_gene_1245 | |
| variant_nucleotide_1245 | |
| variant_protein_1245 | |
| variant_gene_1246 | |
| variant_nucleotide_1246 | |
| variant_protein_1246 | |
| variant_gene_1247 | |
| variant_nucleotide_1247 | |
| variant_protein_1247 | |
| variant_gene_1248 | |
| variant_nucleotide_1248 | |
| variant_protein_1248 | |
| variant_gene_1249 | |
| variant_nucleotide_1249 | |
| variant_protein_1249 | |
| variant_gene_1250 | |
| variant_nucleotide_1250 | |
| variant_protein_1250 | |
| variant_gene_1251 | |
| variant_nucleotide_1251 | |
| variant_protein_1251 | |
| variant_gene_1252 | |
| variant_nucleotide_1252 | |
| variant_protein_1252 | |
| variant_gene_1253 | |
| variant_nucleotide_1253 | |
| variant_protein_1253 | |
| variant_gene_1254 | |
| variant_nucleotide_1254 | |
| variant_protein_1254 | |
| variant_gene_1255 | |
| variant_nucleotide_1255 | |
| variant_protein_1255 | |
| variant_gene_1256 | |
| variant_nucleotide_1256 | |
| variant_protein_1256 | |
| variant_gene_1257 | |
| variant_nucleotide_1257 | |
| variant_protein_1257 | |
| variant_gene_1258 | |
| variant_nucleotide_1258 | |
| variant_protein_1258 | |
| variant_gene_1259 | |
| variant_nucleotide_1259 | |
| variant_protein_1259 | |
| variant_gene_1260 | |
| variant_nucleotide_1260 | |
| variant_protein_1260 | |
| variant_gene_1261 | |
| variant_nucleotide_1261 | |
| variant_protein_1261 | |
| variant_gene_1262 | |
| variant_nucleotide_1262 | |
| variant_protein_1262 | |
| variant_gene_1263 | |
| variant_nucleotide_1263 | |
| variant_protein_1263 | |
| variant_gene_1264 | |
| variant_nucleotide_1264 | |
| variant_protein_1264 | |
| variant_gene_1265 | |
| variant_nucleotide_1265 | |
| variant_protein_1265 | |
| variant_gene_1266 | |
| variant_nucleotide_1266 | |
| variant_protein_1266 | |
| variant_gene_1267 | |
| variant_nucleotide_1267 | |
| variant_protein_1267 | |
| variant_gene_1268 | |
| variant_nucleotide_1268 | |
| variant_protein_1268 | |
| variant_gene_1269 | |
| variant_nucleotide_1269 | |
| variant_protein_1269 | |
| variant_gene_1270 | |
| variant_nucleotide_1270 | |
| variant_protein_1270 | |
| variant_gene_1271 | |
| variant_nucleotide_1271 | |
| variant_protein_1271 | |
| variant_gene_1272 | |
| variant_nucleotide_1272 | |
| variant_protein_1272 | |
| variant_gene_1273 | |
| variant_nucleotide_1273 | |
| variant_protein_1273 | |
| variant_gene_1274 | |
| variant_nucleotide_1274 | |
| variant_protein_1274 | |
| variant_gene_1275 | |
| variant_nucleotide_1275 | |
| variant_protein_1275 | |
| variant_gene_1276 | |
| variant_nucleotide_1276 | |
| variant_protein_1276 | |
| variant_gene_1277 | |
| variant_nucleotide_1277 | |
| variant_protein_1277 | |
| variant_gene_1278 | |
| variant_nucleotide_1278 | |
| variant_protein_1278 | |
| variant_gene_1279 | |
| variant_nucleotide_1279 | |
| variant_protein_1279 | |
| variant_gene_1280 | |
| variant_nucleotide_1280 | |
| variant_protein_1280 | |
| variant_gene_1281 | |
| variant_nucleotide_1281 | |
| variant_protein_1281 | |
| variant_gene_1282 | |
| variant_nucleotide_1282 | |
| variant_protein_1282 | |
| variant_gene_1283 | |
| variant_nucleotide_1283 | |
| variant_protein_1283 | |
| variant_gene_1284 | |
| variant_nucleotide_1284 | |
| variant_protein_1284 | |
| variant_gene_1285 | |
| variant_nucleotide_1285 | |
| variant_protein_1285 | |
| variant_gene_1286 | |
| variant_nucleotide_1286 | |
| variant_protein_1286 | |
| variant_gene_1287 | |
| variant_nucleotide_1287 | |
| variant_protein_1287 | |
| variant_gene_1288 | |
| variant_nucleotide_1288 | |
| variant_protein_1288 | |
| variant_gene_1289 | |
| variant_nucleotide_1289 | |
| variant_protein_1289 | |
| variant_gene_1290 | |
| variant_nucleotide_1290 | |
| variant_protein_1290 | |
| variant_gene_1291 | |
| variant_nucleotide_1291 | |
| variant_protein_1291 | |
| variant_gene_1292 | |
| variant_nucleotide_1292 | |
| variant_protein_1292 | |
| variant_gene_1293 | |
| variant_nucleotide_1293 | |
| variant_protein_1293 | |
| variant_gene_1294 | |
| variant_nucleotide_1294 | |
| variant_protein_1294 | |
| variant_gene_1295 | |
| variant_nucleotide_1295 | |
| variant_protein_1295 | |
| variant_gene_1296 | |
| variant_nucleotide_1296 | |
| variant_protein_1296 | |
| variant_gene_1297 | |
| variant_nucleotide_1297 | |
| variant_protein_1297 | |
| variant_gene_1298 | |
| variant_nucleotide_1298 | |
| variant_protein_1298 | |
| variant_gene_1299 | |
| variant_nucleotide_1299 | |
| variant_protein_1299 | |
| variant_gene_1300 | |
| variant_nucleotide_1300 | |
| variant_protein_1300 | |
| variant_gene_1301 | |
| variant_nucleotide_1301 | |
| variant_protein_1301 | |
| variant_gene_1302 | |
| variant_nucleotide_1302 | |
| variant_protein_1302 | |
| variant_gene_1303 | |
| variant_nucleotide_1303 | |
| variant_protein_1303 | |
| variant_gene_1304 | |
| variant_nucleotide_1304 | |
| variant_protein_1304 | |
| variant_gene_1305 | |
| variant_nucleotide_1305 | |
| variant_protein_1305 | |
| variant_gene_1306 | |
| variant_nucleotide_1306 | |
| variant_protein_1306 | |
| variant_gene_1307 | |
| variant_nucleotide_1307 | |
| variant_protein_1307 | |
| variant_gene_1308 | |
| variant_nucleotide_1308 | |
| variant_protein_1308 | |
| variant_gene_1309 | |
| variant_nucleotide_1309 | |
| variant_protein_1309 | |
| variant_gene_1310 | |
| variant_nucleotide_1310 | |
| variant_protein_1310 | |
| variant_gene_1311 | |
| variant_nucleotide_1311 | |
| variant_protein_1311 | |
| variant_gene_1312 | |
| variant_nucleotide_1312 | |
| variant_protein_1312 | |
| variant_gene_1313 | |
| variant_nucleotide_1313 | |
| variant_protein_1313 | |
| variant_gene_1314 | |
| variant_nucleotide_1314 | |
| variant_protein_1314 | |
| variant_gene_1315 | |
| variant_nucleotide_1315 | |
| variant_protein_1315 | |
| variant_gene_1316 | |
| variant_nucleotide_1316 | |
| variant_protein_1316 | |
| variant_gene_1317 | |
| variant_nucleotide_1317 | |
| variant_protein_1317 | |
| variant_gene_1318 | |
| variant_nucleotide_1318 | |
| variant_protein_1318 | |
| variant_gene_1319 | |
| variant_nucleotide_1319 | |
| variant_protein_1319 | |
| variant_gene_1320 | |
| variant_nucleotide_1320 | |
| variant_protein_1320 | |
| variant_gene_1321 | |
| variant_nucleotide_1321 | |
| variant_protein_1321 | |
| variant_gene_1322 | |
| variant_nucleotide_1322 | |
| variant_protein_1322 | |
| variant_gene_1323 | |
| variant_nucleotide_1323 | |
| variant_protein_1323 | |
| variant_gene_1324 | |
| variant_nucleotide_1324 | |
| variant_protein_1324 | |
| variant_gene_1325 | |
| variant_nucleotide_1325 | |
| variant_protein_1325 | |
| variant_gene_1326 | |
| variant_nucleotide_1326 | |
| variant_protein_1326 | |
| variant_gene_1327 | |
| variant_nucleotide_1327 | |
| variant_protein_1327 | |
| variant_gene_1328 | |
| variant_nucleotide_1328 | |
| variant_protein_1328 | |
| variant_gene_1329 | |
| variant_nucleotide_1329 | |
| variant_protein_1329 | |
| variant_gene_1330 | |
| variant_nucleotide_1330 | |
| variant_protein_1330 | |
| variant_gene_1331 | |
| variant_nucleotide_1331 | |
| variant_protein_1331 | |
| variant_gene_1332 | |
| variant_nucleotide_1332 | |
| variant_protein_1332 | |
| variant_gene_1333 | |
| variant_nucleotide_1333 | |
| variant_protein_1333 | |
| variant_gene_1334 | |
| variant_nucleotide_1334 | |
| variant_protein_1334 | |
| variant_gene_1335 | |
| variant_nucleotide_1335 | |
| variant_protein_1335 | |
| variant_gene_1336 | |
| variant_nucleotide_1336 | |
| variant_protein_1336 | |
| variant_gene_1337 | |
| variant_nucleotide_1337 | |
| variant_protein_1337 | |
| variant_gene_1338 | |
| variant_nucleotide_1338 | |
| variant_protein_1338 | |
| variant_gene_1339 | |
| variant_nucleotide_1339 | |
| variant_protein_1339 | |
| variant_gene_1340 | |
| variant_nucleotide_1340 | |
| variant_protein_1340 | |
| variant_gene_1341 | |
| variant_nucleotide_1341 | |
| variant_protein_1341 | |
| variant_gene_1342 | |
| variant_nucleotide_1342 | |
| variant_protein_1342 | |
| variant_gene_1343 | |
| variant_nucleotide_1343 | |
| variant_protein_1343 | |
| variant_gene_1344 | |
| variant_nucleotide_1344 | |
| variant_protein_1344 | |
| variant_gene_1345 | |
| variant_nucleotide_1345 | |
| variant_protein_1345 | |
| variant_gene_1346 | |
| variant_nucleotide_1346 | |
| variant_protein_1346 | |
| variant_gene_1347 | |
| variant_nucleotide_1347 | |
| variant_protein_1347 | |
| variant_gene_1348 | |
| variant_nucleotide_1348 | |
| variant_protein_1348 | |
| variant_gene_1349 | |
| variant_nucleotide_1349 | |
| variant_protein_1349 | |
| variant_gene_1350 | |
| variant_nucleotide_1350 | |
| variant_protein_1350 | |
| variant_gene_1351 | |
| variant_nucleotide_1351 | |
| variant_protein_1351 | |
| variant_gene_1352 | |
| variant_nucleotide_1352 | |
| variant_protein_1352 | |
| variant_gene_1353 | |
| variant_nucleotide_1353 | |
| variant_protein_1353 | |
| variant_gene_1354 | |
| variant_nucleotide_1354 | |
| variant_protein_1354 | |
| variant_gene_1355 | |
| variant_nucleotide_1355 | |
| variant_protein_1355 | |
| variant_gene_1356 | |
| variant_nucleotide_1356 | |
| variant_protein_1356 | |
| variant_gene_1357 | |
| variant_nucleotide_1357 | |
| variant_protein_1357 | |
| variant_gene_1358 | |
| variant_nucleotide_1358 | |
| variant_protein_1358 | |
| variant_gene_1359 | |
| variant_nucleotide_1359 | |
| variant_protein_1359 | |
| variant_gene_1360 | |
| variant_nucleotide_1360 | |
| variant_protein_1360 | |
| variant_gene_1361 | |
| variant_nucleotide_1361 | |
| variant_protein_1361 | |
| variant_gene_1362 | |
| variant_nucleotide_1362 | |
| variant_protein_1362 | |
| variant_gene_1363 | |
| variant_nucleotide_1363 | |
| variant_protein_1363 | |
| variant_gene_1364 | |
| variant_nucleotide_1364 | |
| variant_protein_1364 | |
| variant_gene_1365 | |
| variant_nucleotide_1365 | |
| variant_protein_1365 | |
| variant_gene_1366 | |
| variant_nucleotide_1366 | |
| variant_protein_1366 | |
| variant_gene_1367 | |
| variant_nucleotide_1367 | |
| variant_protein_1367 | |
| variant_gene_1368 | |
| variant_nucleotide_1368 | |
| variant_protein_1368 | |
| variant_gene_1369 | |
| variant_nucleotide_1369 | |
| variant_protein_1369 | |
| variant_gene_1370 | |
| variant_nucleotide_1370 | |
| variant_protein_1370 | |
| variant_gene_1371 | |
| variant_nucleotide_1371 | |
| variant_protein_1371 | |
| variant_gene_1372 | |
| variant_nucleotide_1372 | |
| variant_protein_1372 | |
| variant_gene_1373 | |
| variant_nucleotide_1373 | |
| variant_protein_1373 | |
| variant_gene_1374 | |
| variant_nucleotide_1374 | |
| variant_protein_1374 | |
| variant_gene_1375 | |
| variant_nucleotide_1375 | |
| variant_protein_1375 | |
| variant_gene_1376 | |
| variant_nucleotide_1376 | |
| variant_protein_1376 | |
| variant_gene_1377 | |
| variant_nucleotide_1377 | |
| variant_protein_1377 | |
| variant_gene_1378 | |
| variant_nucleotide_1378 | |
| variant_protein_1378 | |
| variant_gene_1379 | |
| variant_nucleotide_1379 | |
| variant_protein_1379 | |
| variant_gene_1380 | |
| variant_nucleotide_1380 | |
| variant_protein_1380 | |
| variant_gene_1381 | |
| variant_nucleotide_1381 | |
| variant_protein_1381 | |
| variant_gene_1382 | |
| variant_nucleotide_1382 | |
| variant_protein_1382 | |
| variant_gene_1383 | |
| variant_nucleotide_1383 | |
| variant_protein_1383 | |
| variant_gene_1384 | |
| variant_nucleotide_1384 | |
| variant_protein_1384 | |
| variant_gene_1385 | |
| variant_nucleotide_1385 | |
| variant_protein_1385 | |
| variant_gene_1386 | |
| variant_nucleotide_1386 | |
| variant_protein_1386 | |
| variant_gene_1387 | |
| variant_nucleotide_1387 | |
| variant_protein_1387 | |
| variant_gene_1388 | |
| variant_nucleotide_1388 | |
| variant_protein_1388 | |
| variant_gene_1389 | |
| variant_nucleotide_1389 | |
| variant_protein_1389 | |
| variant_gene_1390 | |
| variant_nucleotide_1390 | |
| variant_protein_1390 | |
| variant_gene_1391 | |
| variant_nucleotide_1391 | |
| variant_protein_1391 | |
| variant_gene_1392 | |
| variant_nucleotide_1392 | |
| variant_protein_1392 | |
| variant_gene_1393 | |
| variant_nucleotide_1393 | |
| variant_protein_1393 | |
| variant_gene_1394 | |
| variant_nucleotide_1394 | |
| variant_protein_1394 | |
| variant_gene_1395 | |
| variant_nucleotide_1395 | |
| variant_protein_1395 | |
| variant_gene_1396 | |
| variant_nucleotide_1396 | |
| variant_protein_1396 | |
| variant_gene_1397 | |
| variant_nucleotide_1397 | |
| variant_protein_1397 | |
| variant_gene_1398 | |
| variant_nucleotide_1398 | |
| variant_protein_1398 | |
| variant_gene_1399 | |
| variant_nucleotide_1399 | |
| variant_protein_1399 | |
| variant_gene_1400 | |
| variant_nucleotide_1400 | |
| variant_protein_1400 | |
| variant_gene_1401 | |
| variant_nucleotide_1401 | |
| variant_protein_1401 | |
| variant_gene_1402 | |
| variant_nucleotide_1402 | |
| variant_protein_1402 | |
| variant_gene_1403 | |
| variant_nucleotide_1403 | |
| variant_protein_1403 | |
| variant_gene_1404 | |
| variant_nucleotide_1404 | |
| variant_protein_1404 | |
| variant_gene_1405 | |
| variant_nucleotide_1405 | |
| variant_protein_1405 | |
| variant_gene_1406 | |
| variant_nucleotide_1406 | |
| variant_protein_1406 | |
| variant_gene_1407 | |
| variant_nucleotide_1407 | |
| variant_protein_1407 | |
| variant_gene_1408 | |
| variant_nucleotide_1408 | |
| variant_protein_1408 | |
| variant_gene_1409 | |
| variant_nucleotide_1409 | |
| variant_protein_1409 | |
| variant_gene_1410 | |
| variant_nucleotide_1410 | |
| variant_protein_1410 | |
| variant_gene_1411 | |
| variant_nucleotide_1411 | |
| variant_protein_1411 | |
| variant_gene_1412 | |
| variant_nucleotide_1412 | |
| variant_protein_1412 | |
| variant_gene_1413 | |
| variant_nucleotide_1413 | |
| variant_protein_1413 | |
| variant_gene_1414 | |
| variant_nucleotide_1414 | |
| variant_protein_1414 | |
| variant_gene_1415 | |
| variant_nucleotide_1415 | |
| variant_protein_1415 | |
| variant_gene_1416 | |
| variant_nucleotide_1416 | |
| variant_protein_1416 | |
| variant_gene_1417 | |
| variant_nucleotide_1417 | |
| variant_protein_1417 | |
| variant_gene_1418 | |
| variant_nucleotide_1418 | |
| variant_protein_1418 | |
| variant_gene_1419 | |
| variant_nucleotide_1419 | |
| variant_protein_1419 | |
| variant_gene_1420 | |
| variant_nucleotide_1420 | |
| variant_protein_1420 | |
| variant_gene_1421 | |
| variant_nucleotide_1421 | |
| variant_protein_1421 | |
| variant_gene_1422 | |
| variant_nucleotide_1422 | |
| variant_protein_1422 | |
| variant_gene_1423 | |
| variant_nucleotide_1423 | |
| variant_protein_1423 | |
| variant_gene_1424 | |
| variant_nucleotide_1424 | |
| variant_protein_1424 | |
| variant_gene_1425 | |
| variant_nucleotide_1425 | |
| variant_protein_1425 | |
| variant_gene_1426 | |
| variant_nucleotide_1426 | |
| variant_protein_1426 | |
| variant_gene_1427 | |
| variant_nucleotide_1427 | |
| variant_protein_1427 | |
| variant_gene_1428 | |
| variant_nucleotide_1428 | |
| variant_protein_1428 | |
| variant_gene_1429 | |
| variant_nucleotide_1429 | |
| variant_protein_1429 | |
| variant_gene_1430 | |
| variant_nucleotide_1430 | |
| variant_protein_1430 | |
| variant_gene_1431 | |
| variant_nucleotide_1431 | |
| variant_protein_1431 | |
| variant_gene_1432 | |
| variant_nucleotide_1432 | |
| variant_protein_1432 | |
| variant_gene_1433 | |
| variant_nucleotide_1433 | |
| variant_protein_1433 | |
| variant_gene_1434 | |
| variant_nucleotide_1434 | |
| variant_protein_1434 | |
| variant_gene_1435 | |
| variant_nucleotide_1435 | |
| variant_protein_1435 | |
| variant_gene_1436 | |
| variant_nucleotide_1436 | |
| variant_protein_1436 | |
| variant_gene_1437 | |
| variant_nucleotide_1437 | |
| variant_protein_1437 | |
| variant_gene_1438 | |
| variant_nucleotide_1438 | |
| variant_protein_1438 | |
| variant_gene_1439 | |
| variant_nucleotide_1439 | |
| variant_protein_1439 | |
| variant_gene_1440 | |
| variant_nucleotide_1440 | |
| variant_protein_1440 | |
| variant_gene_1441 | |
| variant_nucleotide_1441 | |
| variant_protein_1441 | |
| variant_gene_1442 | |
| variant_nucleotide_1442 | |
| variant_protein_1442 | |
| variant_gene_1443 | |
| variant_nucleotide_1443 | |
| variant_protein_1443 | |
| variant_gene_1444 | |
| variant_nucleotide_1444 | |
| variant_protein_1444 | |
| variant_gene_1445 | |
| variant_nucleotide_1445 | |
| variant_protein_1445 | |
| variant_gene_1446 | |
| variant_nucleotide_1446 | |
| variant_protein_1446 | |
| variant_gene_1447 | |
| variant_nucleotide_1447 | |
| variant_protein_1447 | |
| variant_gene_1448 | |
| variant_nucleotide_1448 | |
| variant_protein_1448 | |
| variant_gene_1449 | |
| variant_nucleotide_1449 | |
| variant_protein_1449 | |
| variant_gene_1450 | |
| variant_nucleotide_1450 | |
| variant_protein_1450 | |
| variant_gene_1451 | |
| variant_nucleotide_1451 | |
| variant_protein_1451 | |
| variant_gene_1452 | |
| variant_nucleotide_1452 | |
| variant_protein_1452 | |
| variant_gene_1453 | |
| variant_nucleotide_1453 | |
| variant_protein_1453 | |
| variant_gene_1454 | |
| variant_nucleotide_1454 | |
| variant_protein_1454 | |
| variant_gene_1455 | |
| variant_nucleotide_1455 | |
| variant_protein_1455 | |
| variant_gene_1456 | |
| variant_nucleotide_1456 | |
| variant_protein_1456 | |
| variant_gene_1457 | |
| variant_nucleotide_1457 | |
| variant_protein_1457 | |
| variant_gene_1458 | |
| variant_nucleotide_1458 | |
| variant_protein_1458 | |
| variant_gene_1459 | |
| variant_nucleotide_1459 | |
| variant_protein_1459 | |
| variant_gene_1460 | |
| variant_nucleotide_1460 | |
| variant_protein_1460 | |
| variant_gene_1461 | |
| variant_nucleotide_1461 | |
| variant_protein_1461 | |
| variant_gene_1462 | |
| variant_nucleotide_1462 | |
| variant_protein_1462 | |
| variant_gene_1463 | |
| variant_nucleotide_1463 | |
| variant_protein_1463 | |
| variant_gene_1464 | |
| variant_nucleotide_1464 | |
| variant_protein_1464 | |
| variant_gene_1465 | |
| variant_nucleotide_1465 | |
| variant_protein_1465 | |
| variant_gene_1466 | |
| variant_nucleotide_1466 | |
| variant_protein_1466 | |
| variant_gene_1467 | |
| variant_nucleotide_1467 | |
| variant_protein_1467 | |
| variant_gene_1468 | |
| variant_nucleotide_1468 | |
| variant_protein_1468 | |
| variant_gene_1469 | |
| variant_nucleotide_1469 | |
| variant_protein_1469 | |
| variant_gene_1470 | |
| variant_nucleotide_1470 | |
| variant_protein_1470 | |
| variant_gene_1471 | |
| variant_nucleotide_1471 | |
| variant_protein_1471 | |
| variant_gene_1472 | |
| variant_nucleotide_1472 | |
| variant_protein_1472 | |
| variant_gene_1473 | |
| variant_nucleotide_1473 | |
| variant_protein_1473 | |
| variant_gene_1474 | |
| variant_nucleotide_1474 | |
| variant_protein_1474 | |
| variant_gene_1475 | |
| variant_nucleotide_1475 | |
| variant_protein_1475 | |
| variant_gene_1476 | |
| variant_nucleotide_1476 | |
| variant_protein_1476 | |
| variant_gene_1477 | |
| variant_nucleotide_1477 | |
| variant_protein_1477 | |
| variant_gene_1478 | |
| variant_nucleotide_1478 | |
| variant_protein_1478 | |
| variant_gene_1479 | |
| variant_nucleotide_1479 | |
| variant_protein_1479 | |
| variant_gene_1480 | |
| variant_nucleotide_1480 | |
| variant_protein_1480 | |
| variant_gene_1481 | |
| variant_nucleotide_1481 | |
| variant_protein_1481 | |
| variant_gene_1482 | |
| variant_nucleotide_1482 | |
| variant_protein_1482 | |
| variant_gene_1483 | |
| variant_nucleotide_1483 | |
| variant_protein_1483 | |
| variant_gene_1484 | |
| variant_nucleotide_1484 | |
| variant_protein_1484 | |
| variant_gene_1485 | |
| variant_nucleotide_1485 | |
| variant_protein_1485 | |
| variant_gene_1486 | |
| variant_nucleotide_1486 | |
| variant_protein_1486 | |
| variant_gene_1487 | |
| variant_nucleotide_1487 | |
| variant_protein_1487 | |
| variant_gene_1488 | |
| variant_nucleotide_1488 | |
| variant_protein_1488 | |
| variant_gene_1489 | |
| variant_nucleotide_1489 | |
| variant_protein_1489 | |
| variant_gene_1490 | |
| variant_nucleotide_1490 | |
| variant_protein_1490 | |
| variant_gene_1491 | |
| variant_nucleotide_1491 | |
| variant_protein_1491 | |
| variant_gene_1492 | |
| variant_nucleotide_1492 | |
| variant_protein_1492 | |
| variant_gene_1493 | |
| variant_nucleotide_1493 | |
| variant_protein_1493 | |
| variant_gene_1494 | |
| variant_nucleotide_1494 | |
| variant_protein_1494 | |
| variant_gene_1495 | |
| variant_nucleotide_1495 | |
| variant_protein_1495 | |
| variant_gene_1496 | |
| variant_nucleotide_1496 | |
| variant_protein_1496 | |
| variant_gene_1497 | |
| variant_nucleotide_1497 | |
| variant_protein_1497 | |
| variant_gene_1498 | |
| variant_nucleotide_1498 | |
| variant_protein_1498 | |
| variant_gene_1499 | |
| variant_nucleotide_1499 | |
| variant_protein_1499 | |
| variant_gene_1500 | |
| variant_nucleotide_1500 | |
| variant_protein_1500 | |
| variant_gene_1501 | |
| variant_nucleotide_1501 | |
| variant_protein_1501 | |
| variant_gene_perc_cfdna_1501 | |
| variant_gene_1502 | |
| variant_nucleotide_1502 | |
| variant_protein_1502 | |
| variant_gene_perc_cfdna_1502 | |
| variant_gene_1503 | |
| variant_nucleotide_1503 | |
| variant_protein_1503 | |
| variant_gene_perc_cfdna_1503 | |
| variant_gene_1504 | |
| variant_nucleotide_1504 | |
| variant_protein_1504 | |
| variant_gene_perc_cfdna_1504 | |
| variant_gene_1505 | |
| variant_nucleotide_1505 | |
| variant_protein_1505 | |
| variant_gene_perc_cfdna_1505 | |
| variant_gene_1506 | |
| variant_nucleotide_1506 | |
| variant_protein_1506 | |
| variant_gene_perc_cfdna_1506 | |
| variant_gene_1507 | |
| variant_nucleotide_1507 | |
| variant_protein_1507 | |
| variant_gene_perc_cfdna_1507 | |
| variant_gene_1508 | |
| variant_nucleotide_1508 | |
| variant_protein_1508 | |
| variant_gene_perc_cfdna_1508 | |
| variant_gene_1509 | |
| variant_nucleotide_1509 | |
| variant_protein_1509 | |
| variant_gene_perc_cfdna_1509 | |
| variant_gene_1510 | |
| variant_nucleotide_1510 | |
| variant_protein_1510 | |
| variant_gene_perc_cfdna_1510 | |
| variant_gene_1511 | |
| variant_nucleotide_1511 | |
| variant_protein_1511 | |
| variant_gene_perc_cfdna_1511 | |
| variant_gene_1512 | |
| variant_nucleotide_1512 | |
| variant_protein_1512 | |
| variant_gene_perc_cfdna_1512 | |
| variant_gene_1513 | |
| variant_nucleotide_1513 | |
| variant_protein_1513 | |
| variant_gene_perc_cfdna_1513 | |
| variant_gene_1514 | |
| variant_nucleotide_1514 | |
| variant_protein_1514 | |
| variant_gene_perc_cfdna_1514 | |
| variant_gene_1515 | |
| variant_nucleotide_1515 | |
| variant_protein_1515 | |
| variant_gene_perc_cfdna_1515 | |
| variant_gene_1516 | |
| variant_nucleotide_1516 | |
| variant_protein_1516 | |
| variant_gene_perc_cfdna_1516 | |
| variant_gene_1517 | |
| variant_nucleotide_1517 | |
| variant_protein_1517 | |
| variant_gene_perc_cfdna_1517 | |
| variant_gene_1518 | |
| variant_nucleotide_1518 | |
| variant_protein_1518 | |
| variant_gene_perc_cfdna_1518 | |
| variant_gene_1519 | |
| variant_nucleotide_1519 | |
| variant_protein_1519 | |
| variant_gene_perc_cfdna_1519 | |
| variant_gene_1520 | |
| variant_nucleotide_1520 | |
| variant_protein_1520 | |
| variant_gene_perc_cfdna_1520 | |
| variant_gene_1521 | |
| variant_nucleotide_1521 | |
| variant_protein_1521 | |
| variant_gene_perc_cfdna_1521 | |
| variant_gene_1522 | |
| variant_nucleotide_1522 | |
| variant_protein_1522 | |
| variant_gene_perc_cfdna_1522 | |
| variant_gene_1523 | |
| variant_nucleotide_1523 | |
| variant_protein_1523 | |
| variant_gene_perc_cfdna_1523 | |
| variant_gene_1524 | |
| variant_nucleotide_1524 | |
| variant_protein_1524 | |
| variant_gene_perc_cfdna_1524 | |
| variant_gene_1525 | |
| variant_nucleotide_1525 | |
| variant_protein_1525 | |
| variant_gene_perc_cfdna_1525 | |
| variant_gene_1526 | |
| variant_nucleotide_1526 | |
| variant_protein_1526 | |
| variant_gene_perc_cfdna_1526 | |
| variant_gene_1527 | |
| variant_nucleotide_1527 | |
| variant_protein_1527 | |
| variant_gene_perc_cfdna_1527 | |
| variant_gene_1528 | |
| variant_nucleotide_1528 | |
| variant_protein_1528 | |
| variant_gene_perc_cfdna_1528 | |
| variant_gene_1529 | |
| variant_nucleotide_1529 | |
| variant_protein_1529 | |
| variant_gene_perc_cfdna_1529 | |
| variant_gene_1530 | |
| variant_nucleotide_1530 | |
| variant_protein_1530 | |
| variant_gene_perc_cfdna_1530 | |
| variant_gene_1531 | |
| variant_nucleotide_1531 | |
| variant_protein_1531 | |
| variant_gene_perc_cfdna_1531 | |
| variant_gene_1532 | |
| variant_nucleotide_1532 | |
| variant_protein_1532 | |
| variant_gene_perc_cfdna_1532 | |
| variant_gene_1533 | |
| variant_nucleotide_1533 | |
| variant_protein_1533 | |
| variant_gene_perc_cfdna_1533 | |
| variant_gene_1534 | |
| variant_nucleotide_1534 | |
| variant_protein_1534 | |
| variant_gene_perc_cfdna_1534 | |
| variant_gene_1535 | |
| variant_nucleotide_1535 | |
| variant_protein_1535 | |
| variant_gene_perc_cfdna_1535 | |
| variant_gene_1536 | |
| variant_nucleotide_1536 | |
| variant_protein_1536 | |
| variant_gene_perc_cfdna_1536 | |
| variant_gene_1537 | |
| variant_nucleotide_1537 | |
| variant_protein_1537 | |
| variant_gene_perc_cfdna_1537 | |
| variant_gene_1538 | |
| variant_nucleotide_1538 | |
| variant_protein_1538 | |
| variant_gene_perc_cfdna_1538 | |
| variant_gene_1539 | |
| variant_nucleotide_1539 | |
| variant_protein_1539 | |
| variant_gene_perc_cfdna_1539 | |
| variant_gene_1540 | |
| variant_nucleotide_1540 | |
| variant_protein_1540 | |
| variant_gene_perc_cfdna_1540 | |
| variant_gene_1541 | |
| variant_nucleotide_1541 | |
| variant_protein_1541 | |
| variant_gene_perc_cfdna_1541 | |
| variant_gene_1542 | |
| variant_nucleotide_1542 | |
| variant_protein_1542 | |
| variant_gene_perc_cfdna_1542 | |
| variant_gene_1543 | |
| variant_nucleotide_1543 | |
| variant_protein_1543 | |
| variant_gene_perc_cfdna_1543 | |
| variant_gene_1544 | |
| variant_nucleotide_1544 | |
| variant_protein_1544 | |
| variant_gene_perc_cfdna_1544 | |
| variant_gene_1545 | |
| variant_nucleotide_1545 | |
| variant_protein_1545 | |
| variant_gene_perc_cfdna_1545 | |
| variant_gene_1546 | |
| variant_nucleotide_1546 | |
| variant_protein_1546 | |
| variant_gene_perc_cfdna_1546 | |
| variant_gene_1547 | |
| variant_nucleotide_1547 | |
| variant_protein_1547 | |
| variant_gene_perc_cfdna_1547 | |
| variant_gene_1548 | |
| variant_nucleotide_1548 | |
| variant_protein_1548 | |
| variant_gene_perc_cfdna_1548 | |
| variant_gene_1549 | |
| variant_nucleotide_1549 | |
| variant_protein_1549 | |
| variant_gene_perc_cfdna_1549 | |
| variant_gene_1550 | |
| variant_nucleotide_1550 | |
| variant_protein_1550 | |
| variant_gene_perc_cfdna_1550 | |
| variant_gene_1551 | |
| variant_nucleotide_1551 | |
| variant_protein_1551 | |
| variant_gene_perc_cfdna_1551 | |
| variant_gene_1552 | |
| variant_nucleotide_1552 | |
| variant_protein_1552 | |
| variant_gene_perc_cfdna_1552 | |
| variant_gene_1553 | |
| variant_nucleotide_1553 | |
| variant_protein_1553 | |
| variant_gene_perc_cfdna_1553 | |
| variant_gene_1554 | |
| variant_nucleotide_1554 | |
| variant_protein_1554 | |
| variant_gene_perc_cfdna_1554 | |
| variant_gene_1555 | |
| variant_nucleotide_1555 | |
| variant_protein_1555 | |
| variant_gene_perc_cfdna_1555 | |
| variant_gene_1556 | |
| variant_nucleotide_1556 | |
| variant_protein_1556 | |
| variant_gene_perc_cfdna_1556 | |
| variant_gene_1557 | |
| variant_nucleotide_1557 | |
| variant_protein_1557 | |
| variant_gene_perc_cfdna_1557 | |
| variant_gene_1558 | |
| variant_nucleotide_1558 | |
| variant_protein_1558 | |
| variant_gene_perc_cfdna_1558 | |
| variant_gene_1559 | |
| variant_nucleotide_1559 | |
| variant_protein_1559 | |
| variant_gene_perc_cfdna_1559 | |
| variant_gene_1560 | |
| variant_nucleotide_1560 | |
| variant_protein_1560 | |
| variant_gene_perc_cfdna_1560 | |
| variant_gene_1561 | |
| variant_nucleotide_1561 | |
| variant_protein_1561 | |
| variant_gene_perc_cfdna_1561 | |
| variant_gene_1562 | |
| variant_nucleotide_1562 | |
| variant_protein_1562 | |
| variant_gene_perc_cfdna_1562 | |
| variant_gene_1563 | |
| variant_nucleotide_1563 | |
| variant_protein_1563 | |
| variant_gene_perc_cfdna_1563 | |
| variant_gene_1564 | |
| variant_nucleotide_1564 | |
| variant_protein_1564 | |
| variant_gene_perc_cfdna_1564 | |
| variant_gene_1565 | |
| variant_nucleotide_1565 | |
| variant_protein_1565 | |
| variant_gene_perc_cfdna_1565 | |
| variant_gene_1566 | |
| variant_nucleotide_1566 | |
| variant_protein_1566 | |
| variant_gene_perc_cfdna_1566 | |
| variant_gene_1567 | |
| variant_nucleotide_1567 | |
| variant_protein_1567 | |
| variant_gene_perc_cfdna_1567 | |
| variant_gene_1568 | |
| variant_nucleotide_1568 | |
| variant_protein_1568 | |
| variant_gene_perc_cfdna_1568 | |
| variant_gene_1569 | |
| variant_nucleotide_1569 | |
| variant_protein_1569 | |
| variant_gene_perc_cfdna_1569 | |
| variant_gene_1570 | |
| variant_nucleotide_1570 | |
| variant_protein_1570 | |
| variant_gene_perc_cfdna_1570 | |
| variant_gene_1571 | |
| variant_nucleotide_1571 | |
| variant_protein_1571 | |
| variant_gene_perc_cfdna_1571 | |
| variant_gene_1572 | |
| variant_nucleotide_1572 | |
| variant_protein_1572 | |
| variant_gene_perc_cfdna_1572 | |
| variant_gene_1573 | |
| variant_nucleotide_1573 | |
| variant_protein_1573 | |
| variant_gene_perc_cfdna_1573 | |
| variant_gene_1574 | |
| variant_nucleotide_1574 | |
| variant_protein_1574 | |
| variant_gene_perc_cfdna_1574 | |
| variant_gene_1575 | |
| variant_nucleotide_1575 | |
| variant_protein_1575 | |
| variant_gene_perc_cfdna_1575 | |
| variant_gene_1576 | |
| variant_nucleotide_1576 | |
| variant_protein_1576 | |
| variant_gene_perc_cfdna_1576 | |
| variant_gene_1577 | |
| variant_nucleotide_1577 | |
| variant_protein_1577 | |
| variant_gene_perc_cfdna_1577 | |
| variant_gene_1578 | |
| variant_nucleotide_1578 | |
| variant_protein_1578 | |
| variant_gene_perc_cfdna_1578 | |
| variant_gene_1579 | |
| variant_nucleotide_1579 | |
| variant_protein_1579 | |
| variant_gene_perc_cfdna_1579 | |
| variant_gene_1580 | |
| variant_nucleotide_1580 | |
| variant_protein_1580 | |
| variant_gene_perc_cfdna_1580 | |
| variant_gene_1581 | |
| variant_nucleotide_1581 | |
| variant_protein_1581 | |
| variant_gene_perc_cfdna_1581 | |
| variant_gene_1582 | |
| variant_nucleotide_1582 | |
| variant_protein_1582 | |
| variant_gene_perc_cfdna_1582 | |
| variant_gene_1583 | |
| variant_nucleotide_1583 | |
| variant_protein_1583 | |
| variant_gene_perc_cfdna_1583 | |
| variant_gene_1584 | |
| variant_nucleotide_1584 | |
| variant_protein_1584 | |
| variant_gene_perc_cfdna_1584 | |
| variant_gene_1585 | |
| variant_nucleotide_1585 | |
| variant_protein_1585 | |
| variant_gene_perc_cfdna_1585 | |
| variant_gene_1586 | |
| variant_nucleotide_1586 | |
| variant_protein_1586 | |
| variant_gene_perc_cfdna_1586 | |
| variant_gene_1587 | |
| variant_nucleotide_1587 | |
| variant_protein_1587 | |
| variant_gene_perc_cfdna_1587 | |
| variant_gene_1588 | |
| variant_nucleotide_1588 | |
| variant_protein_1588 | |
| variant_gene_perc_cfdna_1588 | |
| variant_gene_1589 | |
| variant_nucleotide_1589 | |
| variant_protein_1589 | |
| variant_gene_perc_cfdna_1589 | |
| variant_gene_1590 | |
| variant_nucleotide_1590 | |
| variant_protein_1590 | |
| variant_gene_perc_cfdna_1590 | |
| variant_gene_1591 | |
| variant_nucleotide_1591 | |
| variant_protein_1591 | |
| variant_gene_perc_cfdna_1591 | |
| variant_gene_1592 | |
| variant_nucleotide_1592 | |
| variant_protein_1592 | |
| variant_gene_perc_cfdna_1592 | |
| variant_gene_1593 | |
| variant_nucleotide_1593 | |
| variant_protein_1593 | |
| variant_gene_perc_cfdna_1593 | |
| variant_gene_1594 | |
| variant_nucleotide_1594 | |
| variant_protein_1594 | |
| variant_gene_perc_cfdna_1594 | |
| variant_gene_1595 | |
| variant_nucleotide_1595 | |
| variant_protein_1595 | |
| variant_gene_perc_cfdna_1595 | |
| variant_gene_1596 | |
| variant_nucleotide_1596 | |
| variant_protein_1596 | |
| variant_gene_perc_cfdna_1596 | |
| variant_gene_1597 | |
| variant_nucleotide_1597 | |
| variant_protein_1597 | |
| variant_gene_perc_cfdna_1597 | |
| variant_gene_1598 | |
| variant_nucleotide_1598 | |
| variant_protein_1598 | |
| variant_gene_perc_cfdna_1598 | |
| variant_gene_1599 | |
| variant_nucleotide_1599 | |
| variant_protein_1599 | |
| variant_gene_perc_cfdna_1599 | |
| variant_gene_1600 | |
| variant_nucleotide_1600 | |
| variant_protein_1600 | |
| variant_gene_perc_cfdna_1600 | |
| cnv_number | 0 |
| cnv_gene_1 | |
| cnv_gain_or_loss_1 | |
| cnv_gene_2 | |
| cnv_gain_or_loss_2 | |
| cnv_gene_3 | |
| cnv_gain_or_loss_3 | |
| cnv_gene_4 | |
| cnv_gain_or_loss_4 | |
| cnv_gene_5 | |
| cnv_gain_or_loss_5 | |
| cnv_gene_6 | |
| cnv_gain_or_loss_6 | |
| cnv_gene_7 | |
| cnv_gain_or_loss_7 | |
| cnv_gene_8 | |
| cnv_gain_or_loss_8 | |
| cnv_gene_9 | |
| cnv_gain_or_loss_9 | |
| cnv_gene_10 | |
| cnv_gain_or_loss_10 | |
| cnv_gene_11 | |
| cnv_gain_or_loss_11 | |
| cnv_gene_12 | |
| cnv_gain_or_loss_12 | |
| cnv_gene_13 | |
| cnv_gain_or_loss_13 | |
| cnv_gene_14 | |
| cnv_gain_or_loss_14 | |
| cnv_gene_15 | |
| cnv_gain_or_loss_15 | |
| cnv_gene_16 | |
| cnv_gain_or_loss_16 | |
| cnv_gene_17 | |
| cnv_gain_or_loss_17 | |
| cnv_gene_18 | |
| cnv_gain_or_loss_18 | |
| cnv_gene_19 | |
| cnv_gain_or_loss_19 | |
| cnv_gene_20 | |
| cnv_gain_or_loss_20 | |
| cnv_gene_21 | |
| cnv_gain_or_loss_21 | |
| cnv_gene_22 | |
| cnv_gain_or_loss_22 | |
| cnv_gene_23 | |
| cnv_gain_or_loss_23 | |
| cnv_gene_24 | |
| cnv_gain_or_loss_24 | |
| cnv_gene_25 | |
| cnv_gain_or_loss_25 | |
| cnv_gene_26 | |
| cnv_gain_or_loss_26 | |
| cnv_gene_27 | |
| cnv_gain_or_loss_27 | |
| cnv_gene_28 | |
| cnv_gain_or_loss_28 | |
| cnv_gene_29 | |
| cnv_gain_or_loss_29 | |
| cnv_gene_30 | |
| cnv_gain_or_loss_30 | |
| cnv_gene_31 | |
| cnv_gain_or_loss_31 | |
| cnv_gene_32 | |
| cnv_gain_or_loss_32 | |
| cnv_gene_33 | |
| cnv_gain_or_loss_33 | |
| cnv_gene_34 | |
| cnv_gain_or_loss_34 | |
| cnv_gene_35 | |
| cnv_gain_or_loss_35 | |
| cnv_gene_36 | |
| cnv_gain_or_loss_36 | |
| cnv_gene_37 | |
| cnv_gain_or_loss_37 | |
| cnv_gene_38 | |
| cnv_gain_or_loss_38 | |
| cnv_gene_39 | |
| cnv_gain_or_loss_39 | |
| cnv_gene_40 | |
| cnv_gain_or_loss_40 | |
| cnv_gene_41 | |
| cnv_gain_or_loss_41 | |
| cnv_gene_42 | |
| cnv_gain_or_loss_42 | |
| cnv_gene_43 | |
| cnv_gain_or_loss_43 | |
| cnv_gene_44 | |
| cnv_gain_or_loss_44 | |
| cnv_gene_45 | |
| cnv_gain_or_loss_45 | |
| cnv_gene_46 | |
| cnv_gain_or_loss_46 | |
| cnv_gene_47 | |
| cnv_gain_or_loss_47 | |
| cnv_gene_48 | |
| cnv_gain_or_loss_48 | |
| cnv_gene_49 | |
| cnv_gain_or_loss_49 | |
| cnv_gene_50 | |
| cnv_gain_or_loss_50 | |
| cnv_gene_51 | |
| cnv_gain_or_loss_51 | |
| cnv_gene_52 | |
| cnv_gain_or_loss_52 | |
| cnv_gene_53 | |
| cnv_gain_or_loss_53 | |
| cnv_gene_54 | |
| cnv_gain_or_loss_54 | |
| cnv_gene_55 | |
| cnv_gain_or_loss_55 | |
| cnv_gene_56 | |
| cnv_gain_or_loss_56 | |
| cnv_gene_57 | |
| cnv_gain_or_loss_57 | |
| cnv_gene_58 | |
| cnv_gain_or_loss_58 | |
| cnv_gene_59 | |
| cnv_gain_or_loss_59 | |
| cnv_gene_60 | |
| cnv_gain_or_loss_60 | |
| cnv_gene_61 | |
| cnv_gain_or_loss_61 | |
| cnv_gene_62 | |
| cnv_gain_or_loss_62 | |
| cnv_gene_63 | |
| cnv_gain_or_loss_63 | |
| cnv_gene_64 | |
| cnv_gain_or_loss_64 | |
| cnv_gene_65 | |
| cnv_gain_or_loss_65 | |
| cnv_gene_66 | |
| cnv_gain_or_loss_66 | |
| cnv_gene_67 | |
| cnv_gain_or_loss_67 | |
| cnv_gene_68 | |
| cnv_gain_or_loss_68 | |
| cnv_gene_69 | |
| cnv_gain_or_loss_69 | |
| cnv_gene_70 | |
| cnv_gain_or_loss_70 | |
| cnv_gene_71 | |
| cnv_gain_or_loss_71 | |
| cnv_gene_72 | |
| cnv_gain_or_loss_72 | |
| cnv_gene_73 | |
| cnv_gain_or_loss_73 | |
| cnv_gene_74 | |
| cnv_gain_or_loss_74 | |
| cnv_gene_75 | |
| cnv_gain_or_loss_75 | |
| cnv_gene_76 | |
| cnv_gain_or_loss_76 | |
| cnv_gene_77 | |
| cnv_gain_or_loss_77 | |
| cnv_gene_78 | |
| cnv_gain_or_loss_78 | |
| cnv_gene_79 | |
| cnv_gain_or_loss_79 | |
| cnv_gene_80 | |
| cnv_gain_or_loss_80 | |
| cnv_gene_81 | |
| cnv_gain_or_loss_81 | |
| cnv_gene_82 | |
| cnv_gain_or_loss_82 | |
| cnv_gene_83 | |
| cnv_gain_or_loss_83 | |
| cnv_gene_84 | |
| cnv_gain_or_loss_84 | |
| cnv_gene_85 | |
| cnv_gain_or_loss_85 | |
| cnv_gene_86 | |
| cnv_gain_or_loss_86 | |
| cnv_gene_87 | |
| cnv_gain_or_loss_87 | |
| cnv_gene_88 | |
| cnv_gain_or_loss_88 | |
| cnv_gene_89 | |
| cnv_gain_or_loss_89 | |
| cnv_gene_90 | |
| cnv_gain_or_loss_90 | |
| cnv_gene_91 | |
| cnv_gain_or_loss_91 | |
| cnv_gene_92 | |
| cnv_gain_or_loss_92 | |
| cnv_gene_93 | |
| cnv_gain_or_loss_93 | |
| cnv_gene_94 | |
| cnv_gain_or_loss_94 | |
| cnv_gene_95 | |
| cnv_gain_or_loss_95 | |
| cnv_gene_96 | |
| cnv_gain_or_loss_96 | |
| cnv_gene_97 | |
| cnv_gain_or_loss_97 | |
| cnv_gene_98 | |
| cnv_gain_or_loss_98 | |
| cnv_gene_99 | |
| cnv_gain_or_loss_99 | |
| cnv_gene_100 | |
| cnv_gain_or_loss_100 | |
| cnv_gene_101 | |
| cnv_gain_or_loss_101 | |
| cnv_gene_102 | |
| cnv_gain_or_loss_102 | |
| cnv_gene_103 | |
| cnv_gain_or_loss_103 | |
| cnv_gene_104 | |
| cnv_gain_or_loss_104 | |
| cnv_gene_105 | |
| cnv_gain_or_loss_105 | |
| cnv_gene_106 | |
| cnv_gain_or_loss_106 | |
| cnv_gene_107 | |
| cnv_gain_or_loss_107 | |
| cnv_gene_108 | |
| cnv_gain_or_loss_108 | |
| cnv_gene_109 | |
| cnv_gain_or_loss_109 | |
| cnv_gene_110 | |
| cnv_gain_or_loss_110 | |
| cnv_gene_111 | |
| cnv_gain_or_loss_111 | |
| cnv_gene_112 | |
| cnv_gain_or_loss_112 | |
| cnv_gene_113 | |
| cnv_gain_or_loss_113 | |
| cnv_gene_114 | |
| cnv_gain_or_loss_114 | |
| cnv_gene_115 | |
| cnv_gain_or_loss_115 | |
| cnv_gene_116 | |
| cnv_gain_or_loss_116 | |
| cnv_gene_117 | |
| cnv_gain_or_loss_117 | |
| cnv_gene_118 | |
| cnv_gain_or_loss_118 | |
| cnv_gene_119 | |
| cnv_gain_or_loss_119 | |
| cnv_gene_120 | |
| cnv_gain_or_loss_120 | |
| cnv_gene_121 | |
| cnv_gain_or_loss_121 | |
| cnv_gene_122 | |
| cnv_gain_or_loss_122 | |
| cnv_gene_123 | |
| cnv_gain_or_loss_123 | |
| cnv_gene_124 | |
| cnv_gain_or_loss_124 | |
| cnv_gene_125 | |
| cnv_gain_or_loss_125 | |
| cnv_gene_126 | |
| cnv_gain_or_loss_126 | |
| cnv_gene_127 | |
| cnv_gain_or_loss_127 | |
| cnv_gene_128 | |
| cnv_gain_or_loss_128 | |
| cnv_gene_129 | |
| cnv_gain_or_loss_129 | |
| cnv_gene_130 | |
| cnv_gain_or_loss_130 | |
| cnv_gene_131 | |
| cnv_gain_or_loss_131 | |
| cnv_gene_132 | |
| cnv_gain_or_loss_132 | |
| cnv_gene_133 | |
| cnv_gain_or_loss_133 | |
| cnv_gene_134 | |
| cnv_gain_or_loss_134 | |
| cnv_gene_135 | |
| cnv_gain_or_loss_135 | |
| cnv_gene_136 | |
| cnv_gain_or_loss_136 | |
| cnv_gene_137 | |
| cnv_gain_or_loss_137 | |
| cnv_gene_138 | |
| cnv_gain_or_loss_138 | |
| cnv_gene_139 | |
| cnv_gain_or_loss_139 | |
| cnv_gene_140 | |
| cnv_gain_or_loss_140 | |
| cnv_gene_141 | |
| cnv_gain_or_loss_141 | |
| cnv_gene_142 | |
| cnv_gain_or_loss_142 | |
| cnv_gene_143 | |
| cnv_gain_or_loss_143 | |
| cnv_gene_144 | |
| cnv_gain_or_loss_144 | |
| cnv_gene_145 | |
| cnv_gain_or_loss_145 | |
| cnv_gene_146 | |
| cnv_gain_or_loss_146 | |
| cnv_gene_147 | |
| cnv_gain_or_loss_147 | |
| cnv_gene_148 | |
| cnv_gain_or_loss_148 | |
| cnv_gene_149 | |
| cnv_gain_or_loss_149 | |
| cnv_gene_150 | |
| cnv_gain_or_loss_150 | |
| cnv_gene_151 | |
| cnv_gain_or_loss_151 | |
| cnv_gene_152 | |
| cnv_gain_or_loss_152 | |
| cnv_gene_153 | |
| cnv_gain_or_loss_153 | |
| cnv_gene_154 | |
| cnv_gain_or_loss_154 | |
| cnv_gene_155 | |
| cnv_gain_or_loss_155 | |
| cnv_gene_156 | |
| cnv_gain_or_loss_156 | |
| cnv_gene_157 | |
| cnv_gain_or_loss_157 | |
| cnv_gene_158 | |
| cnv_gain_or_loss_158 | |
| cnv_gene_159 | |
| cnv_gain_or_loss_159 | |
| cnv_gene_160 | |
| cnv_gain_or_loss_160 | |
| cnv_gene_161 | |
| cnv_gain_or_loss_161 | |
| cnv_gene_162 | |
| cnv_gain_or_loss_162 | |
| cnv_gene_163 | |
| cnv_gain_or_loss_163 | |
| cnv_gene_164 | |
| cnv_gain_or_loss_164 | |
| cnv_gene_165 | |
| cnv_gain_or_loss_165 | |
| cnv_gene_166 | |
| cnv_gain_or_loss_166 | |
| cnv_gene_167 | |
| cnv_gain_or_loss_167 | |
| cnv_gene_168 | |
| cnv_gain_or_loss_168 | |
| cnv_gene_169 | |
| cnv_gain_or_loss_169 | |
| cnv_gene_170 | |
| cnv_gain_or_loss_170 | |
| cnv_gene_171 | |
| cnv_gain_or_loss_171 | |
| cnv_gene_172 | |
| cnv_gain_or_loss_172 | |
| cnv_gene_173 | |
| cnv_gain_or_loss_173 | |
| cnv_gene_174 | |
| cnv_gain_or_loss_174 | |
| cnv_gene_175 | |
| cnv_gain_or_loss_175 | |
| cnv_gene_176 | |
| cnv_gain_or_loss_176 | |
| cnv_gene_177 | |
| cnv_gain_or_loss_177 | |
| cnv_gene_178 | |
| cnv_gain_or_loss_178 | |
| cnv_gene_179 | |
| cnv_gain_or_loss_179 | |
| cnv_gene_180 | |
| cnv_gain_or_loss_180 | |
| cnv_gene_181 | |
| cnv_gain_or_loss_181 | |
| cnv_gene_182 | |
| cnv_gain_or_loss_182 | |
| cnv_gene_183 | |
| cnv_gain_or_loss_183 | |
| cnv_gene_184 | |
| cnv_gain_or_loss_184 | |
| cnv_gene_185 | |
| cnv_gain_or_loss_185 | |
| cnv_gene_186 | |
| cnv_gain_or_loss_186 | |
| cnv_gene_187 | |
| cnv_gain_or_loss_187 | |
| cnv_gene_188 | |
| cnv_gain_or_loss_188 | |
| cnv_gene_189 | |
| cnv_gain_or_loss_189 | |
| cnv_gene_190 | |
| cnv_gain_or_loss_190 | |
| cnv_gene_191 | |
| cnv_gain_or_loss_191 | |
| cnv_gene_192 | |
| cnv_gain_or_loss_192 | |
| cnv_gene_193 | |
| cnv_gain_or_loss_193 | |
| cnv_gene_194 | |
| cnv_gain_or_loss_194 | |
| cnv_gene_195 | |
| cnv_gain_or_loss_195 | |
| cnv_gene_196 | |
| cnv_gain_or_loss_196 | |
| cnv_gene_197 | |
| cnv_gain_or_loss_197 | |
| cnv_gene_198 | |
| cnv_gain_or_loss_198 | |
| cnv_gene_199 | |
| cnv_gain_or_loss_199 | |
| cnv_gene_200 | |
| cnv_gain_or_loss_200 | |
| cnv_gene_201 | |
| cnv_gain_or_loss_201 | |
| cnv_gene_202 | |
| cnv_gain_or_loss_202 | |
| cnv_gene_203 | |
| cnv_gain_or_loss_203 | |
| cnv_gene_204 | |
| cnv_gain_or_loss_204 | |
| cnv_gene_205 | |
| cnv_gain_or_loss_205 | |
| cnv_gene_206 | |
| cnv_gain_or_loss_206 | |
| cnv_gene_207 | |
| cnv_gain_or_loss_207 | |
| cnv_gene_208 | |
| cnv_gain_or_loss_208 | |
| cnv_gene_209 | |
| cnv_gain_or_loss_209 | |
| cnv_gene_210 | |
| cnv_gain_or_loss_210 | |
| cnv_gene_211 | |
| cnv_gain_or_loss_211 | |
| cnv_gene_212 | |
| cnv_gain_or_loss_212 | |
| cnv_gene_213 | |
| cnv_gain_or_loss_213 | |
| cnv_gene_214 | |
| cnv_gain_or_loss_214 | |
| cnv_gene_215 | |
| cnv_gain_or_loss_215 | |
| cnv_gene_216 | |
| cnv_gain_or_loss_216 | |
| cnv_gene_217 | |
| cnv_gain_or_loss_217 | |
| cnv_gene_218 | |
| cnv_gain_or_loss_218 | |
| cnv_gene_219 | |
| cnv_gain_or_loss_219 | |
| cnv_gene_220 | |
| cnv_gain_or_loss_220 | |
| cnv_gene_221 | |
| cnv_gain_or_loss_221 | |
| cnv_gene_222 | |
| cnv_gain_or_loss_222 | |
| cnv_gene_223 | |
| cnv_gain_or_loss_223 | |
| cnv_gene_224 | |
| cnv_gain_or_loss_224 | |
| cnv_gene_225 | |
| cnv_gain_or_loss_225 | |
| cnv_gene_226 | |
| cnv_gain_or_loss_226 | |
| cnv_gene_227 | |
| cnv_gain_or_loss_227 | |
| cnv_gene_228 | |
| cnv_gain_or_loss_228 | |
| cnv_gene_229 | |
| cnv_gain_or_loss_229 | |
| cnv_gene_230 | |
| cnv_gain_or_loss_230 | |
| cnv_gene_231 | |
| cnv_gain_or_loss_231 | |
| cnv_gene_232 | |
| cnv_gain_or_loss_232 | |
| cnv_gene_233 | |
| cnv_gain_or_loss_233 | |
| cnv_gene_234 | |
| cnv_gain_or_loss_234 | |
| cnv_gene_235 | |
| cnv_gain_or_loss_235 | |
| cnv_gene_236 | |
| cnv_gain_or_loss_236 | |
| cnv_gene_237 | |
| cnv_gain_or_loss_237 | |
| cnv_gene_238 | |
| cnv_gain_or_loss_238 | |
| cnv_gene_239 | |
| cnv_gain_or_loss_239 | |
| cnv_gene_240 | |
| cnv_gain_or_loss_240 | |
| cnv_gene_241 | |
| cnv_gain_or_loss_241 | |
| cnv_gene_242 | |
| cnv_gain_or_loss_242 | |
| cnv_gene_243 | |
| cnv_gain_or_loss_243 | |
| cnv_gene_244 | |
| cnv_gain_or_loss_244 | |
| cnv_gene_245 | |
| cnv_gain_or_loss_245 | |
| cnv_gene_246 | |
| cnv_gain_or_loss_246 | |
| cnv_gene_247 | |
| cnv_gain_or_loss_247 | |
| cnv_gene_248 | |
| cnv_gain_or_loss_248 | |
| cnv_gene_249 | |
| cnv_gain_or_loss_249 | |
| cnv_gene_250 | |
| cnv_gain_or_loss_250 | |
| cnv_gene_251 | |
| cnv_gain_or_loss_251 | |
| cnv_gene_252 | |
| cnv_gain_or_loss_252 | |
| cnv_gene_253 | |
| cnv_gain_or_loss_253 | |
| cnv_gene_254 | |
| cnv_gain_or_loss_254 | |
| cnv_gene_255 | |
| cnv_gain_or_loss_255 | |
| cnv_gene_256 | |
| cnv_gain_or_loss_256 | |
| cnv_gene_257 | |
| cnv_gain_or_loss_257 | |
| cnv_gene_258 | |
| cnv_gain_or_loss_258 | |
| cnv_gene_259 | |
| cnv_gain_or_loss_259 | |
| cnv_gene_260 | |
| cnv_gain_or_loss_260 | |
| cnv_gene_261 | |
| cnv_gain_or_loss_261 | |
| cnv_gene_262 | |
| cnv_gain_or_loss_262 | |
| cnv_gene_263 | |
| cnv_gain_or_loss_263 | |
| cnv_gene_264 | |
| cnv_gain_or_loss_264 | |
| cnv_gene_265 | |
| cnv_gain_or_loss_265 | |
| cnv_gene_266 | |
| cnv_gain_or_loss_266 | |
| cnv_gene_267 | |
| cnv_gain_or_loss_267 | |
| cnv_gene_268 | |
| cnv_gain_or_loss_268 | |
| cnv_gene_269 | |
| cnv_gain_or_loss_269 | |
| cnv_gene_270 | |
| cnv_gain_or_loss_270 | |
| cnv_gene_271 | |
| cnv_gain_or_loss_271 | |
| cnv_gene_272 | |
| cnv_gain_or_loss_272 | |
| cnv_gene_273 | |
| cnv_gain_or_loss_273 | |
| cnv_gene_274 | |
| cnv_gain_or_loss_274 | |
| cnv_gene_275 | |
| cnv_gain_or_loss_275 | |
| cnv_gene_276 | |
| cnv_gain_or_loss_276 | |
| cnv_gene_277 | |
| cnv_gain_or_loss_277 | |
| cnv_gene_278 | |
| cnv_gain_or_loss_278 | |
| cnv_gene_279 | |
| cnv_gain_or_loss_279 | |
| cnv_gene_280 | |
| cnv_gain_or_loss_280 | |
| cnv_gene_281 | |
| cnv_gain_or_loss_281 | |
| cnv_gene_282 | |
| cnv_gain_or_loss_282 | |
| cnv_gene_283 | |
| cnv_gain_or_loss_283 | |
| cnv_gene_284 | |
| cnv_gain_or_loss_284 | |
| cnv_gene_285 | |
| cnv_gain_or_loss_285 | |
| cnv_gene_286 | |
| cnv_gain_or_loss_286 | |
| cnv_gene_287 | |
| cnv_gain_or_loss_287 | |
| cnv_gene_288 | |
| cnv_gain_or_loss_288 | |
| cnv_gene_289 | |
| cnv_gain_or_loss_289 | |
| cnv_gene_290 | |
| cnv_gain_or_loss_290 | |
| cnv_gene_291 | |
| cnv_gain_or_loss_291 | |
| cnv_gene_292 | |
| cnv_gain_or_loss_292 | |
| cnv_gene_293 | |
| cnv_gain_or_loss_293 | |
| cnv_gene_294 | |
| cnv_gain_or_loss_294 | |
| cnv_gene_295 | |
| cnv_gain_or_loss_295 | |
| cnv_gene_296 | |
| cnv_gain_or_loss_296 | |
| cnv_gene_297 | |
| cnv_gain_or_loss_297 | |
| cnv_gene_298 | |
| cnv_gain_or_loss_298 | |
| cnv_gene_299 | |
| cnv_gain_or_loss_299 | |
| cnv_gene_300 | |
| cnv_gain_or_loss_300 | |
| cnv_gene_301 | |
| cnv_gain_or_loss_301 | |
| cnv_gene_302 | |
| cnv_gain_or_loss_302 | |
| cnv_gene_303 | |
| cnv_gain_or_loss_303 | |
| cnv_gene_304 | |
| cnv_gain_or_loss_304 | |
| cnv_gene_305 | |
| cnv_gain_or_loss_305 | |
| cnv_gene_306 | |
| cnv_gain_or_loss_306 | |
| cnv_gene_307 | |
| cnv_gain_or_loss_307 | |
| cnv_gene_308 | |
| cnv_gain_or_loss_308 | |
| cnv_gene_309 | |
| cnv_gain_or_loss_309 | |
| cnv_gene_310 | |
| cnv_gain_or_loss_310 | |
| cnv_gene_311 | |
| cnv_gain_or_loss_311 | |
| cnv_gene_312 | |
| cnv_gain_or_loss_312 | |
| cnv_gene_313 | |
| cnv_gain_or_loss_313 | |
| cnv_gene_314 | |
| cnv_gain_or_loss_314 | |
| cnv_gene_315 | |
| cnv_gain_or_loss_315 | |
| cnv_gene_316 | |
| cnv_gain_or_loss_316 | |
| cnv_gene_317 | |
| cnv_gain_or_loss_317 | |
| cnv_gene_318 | |
| cnv_gain_or_loss_318 | |
| cnv_gene_319 | |
| cnv_gain_or_loss_319 | |
| cnv_gene_320 | |
| cnv_gain_or_loss_320 | |
| cnv_gene_321 | |
| cnv_gain_or_loss_321 | |
| cnv_gene_322 | |
| cnv_gain_or_loss_322 | |
| cnv_gene_323 | |
| cnv_gain_or_loss_323 | |
| cnv_gene_324 | |
| cnv_gain_or_loss_324 | |
| cnv_gene_325 | |
| cnv_gain_or_loss_325 | |
| cnv_gene_326 | |
| cnv_gain_or_loss_326 | |
| cnv_gene_327 | |
| cnv_gain_or_loss_327 | |
| cnv_gene_328 | |
| cnv_gain_or_loss_328 | |
| cnv_gene_329 | |
| cnv_gain_or_loss_329 | |
| cnv_gene_330 | |
| cnv_gain_or_loss_330 | |
| cnv_gene_331 | |
| cnv_gain_or_loss_331 | |
| cnv_gene_332 | |
| cnv_gain_or_loss_332 | |
| cnv_gene_333 | |
| cnv_gain_or_loss_333 | |
| cnv_gene_334 | |
| cnv_gain_or_loss_334 | |
| cnv_gene_335 | |
| cnv_gain_or_loss_335 | |
| cnv_gene_336 | |
| cnv_gain_or_loss_336 | |
| cnv_gene_337 | |
| cnv_gain_or_loss_337 | |
| cnv_gene_338 | |
| cnv_gain_or_loss_338 | |
| cnv_gene_339 | |
| cnv_gain_or_loss_339 | |
| cnv_gene_340 | |
| cnv_gain_or_loss_340 | |
| cnv_gene_341 | |
| cnv_gain_or_loss_341 | |
| cnv_gene_342 | |
| cnv_gain_or_loss_342 | |
| cnv_gene_343 | |
| cnv_gain_or_loss_343 | |
| cnv_gene_344 | |
| cnv_gain_or_loss_344 | |
| cnv_gene_345 | |
| cnv_gain_or_loss_345 | |
| cnv_gene_346 | |
| cnv_gain_or_loss_346 | |
| cnv_gene_347 | |
| cnv_gain_or_loss_347 | |
| cnv_gene_348 | |
| cnv_gain_or_loss_348 | |
| cnv_gene_349 | |
| cnv_gain_or_loss_349 | |
| cnv_gene_350 | |
| cnv_gain_or_loss_350 | |
| cnv_gene_351 | |
| cnv_gain_or_loss_351 | |
| cnv_gene_352 | |
| cnv_gain_or_loss_352 | |
| cnv_gene_353 | |
| cnv_gain_or_loss_353 | |
| cnv_gene_354 | |
| cnv_gain_or_loss_354 | |
| cnv_gene_355 | |
| cnv_gain_or_loss_355 | |
| cnv_gene_356 | |
| cnv_gain_or_loss_356 | |
| cnv_gene_357 | |
| cnv_gain_or_loss_357 | |
| cnv_gene_358 | |
| cnv_gain_or_loss_358 | |
| cnv_gene_359 | |
| cnv_gain_or_loss_359 | |
| cnv_gene_360 | |
| cnv_gain_or_loss_360 | |
| cnv_gene_361 | |
| cnv_gain_or_loss_361 | |
| cnv_gene_362 | |
| cnv_gain_or_loss_362 | |
| cnv_gene_363 | |
| cnv_gain_or_loss_363 | |
| cnv_gene_364 | |
| cnv_gain_or_loss_364 | |
| cnv_gene_365 | |
| cnv_gain_or_loss_365 | |
| cnv_gene_366 | |
| cnv_gain_or_loss_366 | |
| cnv_gene_367 | |
| cnv_gain_or_loss_367 | |
| cnv_gene_368 | |
| cnv_gain_or_loss_368 | |
| cnv_gene_369 | |
| cnv_gain_or_loss_369 | |
| cnv_gene_370 | |
| cnv_gain_or_loss_370 | |
| cnv_gene_371 | |
| cnv_gain_or_loss_371 | |
| cnv_gene_372 | |
| cnv_gain_or_loss_372 | |
| cnv_gene_373 | |
| cnv_gain_or_loss_373 | |
| cnv_gene_374 | |
| cnv_gain_or_loss_374 | |
| cnv_gene_375 | |
| cnv_gain_or_loss_375 | |
| cnv_gene_376 | |
| cnv_gain_or_loss_376 | |
| cnv_gene_377 | |
| cnv_gain_or_loss_377 | |
| cnv_gene_378 | |
| cnv_gain_or_loss_378 | |
| cnv_gene_379 | |
| cnv_gain_or_loss_379 | |
| cnv_gene_380 | |
| cnv_gain_or_loss_380 | |
| cnv_gene_381 | |
| cnv_gain_or_loss_381 | |
| cnv_gene_382 | |
| cnv_gain_or_loss_382 | |
| cnv_gene_383 | |
| cnv_gain_or_loss_383 | |
| cnv_gene_384 | |
| cnv_gain_or_loss_384 | |
| cnv_gene_385 | |
| cnv_gain_or_loss_385 | |
| cnv_gene_386 | |
| cnv_gain_or_loss_386 | |
| cnv_gene_387 | |
| cnv_gain_or_loss_387 | |
| cnv_gene_388 | |
| cnv_gain_or_loss_388 | |
| cnv_gene_389 | |
| cnv_gain_or_loss_389 | |
| cnv_gene_390 | |
| cnv_gain_or_loss_390 | |
| cnv_gene_391 | |
| cnv_gain_or_loss_391 | |
| cnv_gene_392 | |
| cnv_gain_or_loss_392 | |
| cnv_gene_393 | |
| cnv_gain_or_loss_393 | |
| cnv_gene_394 | |
| cnv_gain_or_loss_394 | |
| cnv_gene_395 | |
| cnv_gain_or_loss_395 | |
| cnv_gene_396 | |
| cnv_gain_or_loss_396 | |
| cnv_gene_397 | |
| cnv_gain_or_loss_397 | |
| cnv_gene_398 | |
| cnv_gain_or_loss_398 | |
| cnv_gene_399 | |
| cnv_gain_or_loss_399 | |
| cnv_gene_400 | |
| cnv_gain_or_loss_400 | |
| cnv_gene_401 | |
| cnv_gain_or_loss_401 | |
| cnv_gene_402 | |
| cnv_gain_or_loss_402 | |
| cnv_gene_403 | |
| cnv_gain_or_loss_403 | |
| cnv_gene_404 | |
| cnv_gain_or_loss_404 | |
| cnv_gene_405 | |
| cnv_gain_or_loss_405 | |
| cnv_gene_406 | |
| cnv_gain_or_loss_406 | |
| cnv_gene_407 | |
| cnv_gain_or_loss_407 | |
| cnv_gene_408 | |
| cnv_gain_or_loss_408 | |
| cnv_gene_409 | |
| cnv_gain_or_loss_409 | |
| cnv_gene_410 | |
| cnv_gain_or_loss_410 | |
| cnv_gene_411 | |
| cnv_gain_or_loss_411 | |
| cnv_gene_412 | |
| cnv_gain_or_loss_412 | |
| cnv_gene_413 | |
| cnv_gain_or_loss_413 | |
| cnv_gene_414 | |
| cnv_gain_or_loss_414 | |
| cnv_gene_415 | |
| cnv_gain_or_loss_415 | |
| cnv_gene_416 | |
| cnv_gain_or_loss_416 | |
| cnv_gene_417 | |
| cnv_gain_or_loss_417 | |
| cnv_gene_418 | |
| cnv_gain_or_loss_418 | |
| cnv_gene_419 | |
| cnv_gain_or_loss_419 | |
| cnv_gene_420 | |
| cnv_gain_or_loss_420 | |
| cnv_gene_421 | |
| cnv_gain_or_loss_421 | |
| cnv_gene_422 | |
| cnv_gain_or_loss_422 | |
| cnv_gene_423 | |
| cnv_gain_or_loss_423 | |
| cnv_gene_424 | |
| cnv_gain_or_loss_424 | |
| cnv_gene_425 | |
| cnv_gain_or_loss_425 | |
| cnv_gene_426 | |
| cnv_gain_or_loss_426 | |
| cnv_gene_427 | |
| cnv_gain_or_loss_427 | |
| cnv_gene_428 | |
| cnv_gain_or_loss_428 | |
| cnv_gene_429 | |
| cnv_gain_or_loss_429 | |
| cnv_gene_430 | |
| cnv_gain_or_loss_430 | |
| cnv_gene_431 | |
| cnv_gain_or_loss_431 | |
| cnv_gene_432 | |
| cnv_gain_or_loss_432 | |
| cnv_gene_433 | |
| cnv_gain_or_loss_433 | |
| cnv_gene_434 | |
| cnv_gain_or_loss_434 | |
| cnv_gene_435 | |
| cnv_gain_or_loss_435 | |
| cnv_gene_436 | |
| cnv_gain_or_loss_436 | |
| cnv_gene_437 | |
| cnv_gain_or_loss_437 | |
| cnv_gene_438 | |
| cnv_gain_or_loss_438 | |
| cnv_gene_439 | |
| cnv_gain_or_loss_439 | |
| cnv_gene_440 | |
| cnv_gain_or_loss_440 | |
| cnv_gene_441 | |
| cnv_gain_or_loss_441 | |
| cnv_gene_442 | |
| cnv_gain_or_loss_442 | |
| cnv_gene_443 | |
| cnv_gain_or_loss_443 | |
| cnv_gene_444 | |
| cnv_gain_or_loss_444 | |
| cnv_gene_445 | |
| cnv_gain_or_loss_445 | |
| cnv_gene_446 | |
| cnv_gain_or_loss_446 | |
| cnv_gene_447 | |
| cnv_gain_or_loss_447 | |
| cnv_gene_448 | |
| cnv_gain_or_loss_448 | |
| cnv_gene_449 | |
| cnv_gain_or_loss_449 | |
| cnv_gene_450 | |
| cnv_gain_or_loss_450 | |
| cnv_gene_451 | |
| cnv_gain_or_loss_451 | |
| cnv_gene_452 | |
| cnv_gain_or_loss_452 | |
| cnv_gene_453 | |
| cnv_gain_or_loss_453 | |
| cnv_gene_454 | |
| cnv_gain_or_loss_454 | |
| cnv_gene_455 | |
| cnv_gain_or_loss_455 | |
| cnv_gene_456 | |
| cnv_gain_or_loss_456 | |
| cnv_gene_457 | |
| cnv_gain_or_loss_457 | |
| cnv_gene_458 | |
| cnv_gain_or_loss_458 | |
| cnv_gene_459 | |
| cnv_gain_or_loss_459 | |
| cnv_gene_460 | |
| cnv_gain_or_loss_460 | |
| cnv_gene_461 | |
| cnv_gain_or_loss_461 | |
| cnv_gene_462 | |
| cnv_gain_or_loss_462 | |
| cnv_gene_463 | |
| cnv_gain_or_loss_463 | |
| cnv_gene_464 | |
| cnv_gain_or_loss_464 | |
| cnv_gene_465 | |
| cnv_gain_or_loss_465 | |
| cnv_gene_466 | |
| cnv_gain_or_loss_466 | |
| cnv_gene_467 | |
| cnv_gain_or_loss_467 | |
| cnv_gene_468 | |
| cnv_gain_or_loss_468 | |
| cnv_gene_469 | |
| cnv_gain_or_loss_469 | |
| cnv_gene_470 | |
| cnv_gain_or_loss_470 | |
| cnv_gene_471 | |
| cnv_gain_or_loss_471 | |
| cnv_gene_472 | |
| cnv_gain_or_loss_472 | |
| cnv_gene_473 | |
| cnv_gain_or_loss_473 | |
| cnv_gene_474 | |
| cnv_gain_or_loss_474 | |
| cnv_gene_475 | |
| cnv_gain_or_loss_475 | |
| cnv_gene_476 | |
| cnv_gain_or_loss_476 | |
| cnv_gene_477 | |
| cnv_gain_or_loss_477 | |
| cnv_gene_478 | |
| cnv_gain_or_loss_478 | |
| cnv_gene_479 | |
| cnv_gain_or_loss_479 | |
| cnv_gene_480 | |
| cnv_gain_or_loss_480 | |
| cnv_gene_481 | |
| cnv_gain_or_loss_481 | |
| cnv_gene_482 | |
| cnv_gain_or_loss_482 | |
| cnv_gene_483 | |
| cnv_gain_or_loss_483 | |
| cnv_gene_484 | |
| cnv_gain_or_loss_484 | |
| cnv_gene_485 | |
| cnv_gain_or_loss_485 | |
| cnv_gene_486 | |
| cnv_gain_or_loss_486 | |
| cnv_gene_487 | |
| cnv_gain_or_loss_487 | |
| cnv_gene_488 | |
| cnv_gain_or_loss_488 | |
| cnv_gene_489 | |
| cnv_gain_or_loss_489 | |
| cnv_gene_490 | |
| cnv_gain_or_loss_490 | |
| cnv_gene_491 | |
| cnv_gain_or_loss_491 | |
| cnv_gene_492 | |
| cnv_gain_or_loss_492 | |
| cnv_gene_493 | |
| cnv_gain_or_loss_493 | |
| cnv_gene_494 | |
| cnv_gain_or_loss_494 | |
| cnv_gene_495 | |
| cnv_gain_or_loss_495 | |
| cnv_gene_496 | |
| cnv_gain_or_loss_496 | |
| cnv_gene_497 | |
| cnv_gain_or_loss_497 | |
| cnv_gene_498 | |
| cnv_gain_or_loss_498 | |
| cnv_gene_499 | |
| cnv_gain_or_loss_499 | |
| cnv_gene_500 | |
| cnv_gain_or_loss_500 | |
| amplifications_number | 0 |
| amplifications_gene_1 | |
| amplifications_gene_2 | |
| amplifications_gene_3 | |
| amplifications_gene_4 | |
| amplifications_gene_5 | |
| amplifications_gene_6 | |
| amplifications_gene_7 | |
| amplifications_gene_8 | |
| amplifications_gene_9 | |
| amplifications_gene_10 | |
| amplifications_gene_11 | |
| amplifications_gene_12 | |
| amplifications_gene_13 | |
| amplifications_gene_14 | |
| amplifications_gene_15 | |
| amplifications_gene_16 | |
| amplifications_gene_17 | |
| amplifications_gene_18 | |
| amplifications_gene_19 | |
| amplifications_gene_20 | |
| amplifications_gene_21 | |
| amplifications_gene_22 | |
| amplifications_gene_23 | |
| amplifications_gene_24 | |
| amplifications_gene_25 | |
| amplifications_gene_26 | |
| amplifications_gene_27 | |
| amplifications_gene_28 | |
| amplifications_gene_29 | |
| amplifications_gene_30 | |
| amplifications_gene_31 | |
| amplifications_gene_32 | |
| amplifications_gene_33 | |
| amplifications_gene_34 | |
| amplifications_gene_35 | |
| amplifications_gene_36 | |
| amplifications_gene_37 | |
| amplifications_gene_38 | |
| amplifications_gene_39 | |
| amplifications_gene_40 | |
| amplifications_gene_41 | |
| amplifications_gene_42 | |
| amplifications_gene_43 | |
| amplifications_gene_44 | |
| amplifications_gene_45 | |
| amplifications_gene_46 | |
| amplifications_gene_47 | |
| amplifications_gene_48 | |
| amplifications_gene_49 | |
| amplifications_gene_50 | |
| amplifications_gene_51 | |
| amplifications_gene_52 | |
| amplifications_gene_53 | |
| amplifications_gene_54 | |
| amplifications_gene_55 | |
| amplifications_gene_56 | |
| amplifications_gene_57 | |
| amplifications_gene_58 | |
| amplifications_gene_59 | |
| amplifications_gene_60 | |
| amplifications_gene_61 | |
| amplifications_gene_62 | |
| amplifications_gene_63 | |
| amplifications_gene_64 | |
| amplifications_gene_65 | |
| amplifications_gene_66 | |
| amplifications_gene_67 | |
| amplifications_gene_68 | |
| amplifications_gene_69 | |
| amplifications_gene_70 | |
| amplifications_gene_71 | |
| amplifications_gene_72 | |
| amplifications_gene_73 | |
| amplifications_gene_74 | |
| amplifications_gene_75 | |
| amplifications_gene_76 | |
| amplifications_gene_77 | |
| amplifications_gene_78 | |
| amplifications_gene_79 | |
| amplifications_gene_80 | |
| amplifications_gene_81 | |
| amplifications_gene_82 | |
| amplifications_gene_83 | |
| amplifications_gene_84 | |
| amplifications_gene_85 | |
| amplifications_gene_86 | |
| amplifications_gene_87 | |
| amplifications_gene_88 | |
| amplifications_gene_89 | |
| amplifications_gene_90 | |
| amplifications_gene_91 | |
| amplifications_gene_92 | |
| amplifications_gene_93 | |
| amplifications_gene_94 | |
| amplifications_gene_95 | |
| amplifications_gene_96 | |
| amplifications_gene_97 | |
| amplifications_gene_98 | |
| amplifications_gene_99 | |
| amplifications_gene_100 | |
| fused_number | |
| fused_gene_1 | |
| fused_gene_2 | |
| fused_gene_3 | |
| fused_gene_4 | |
| fused_gene_5 | |
| fused_gene_6 | |
| fused_gene_7 | |
| fused_gene_8 | |
| fused_gene_9 | |
| fused_gene_10 | |
| fused_gene_11 | |
| fused_gene_12 | |
| fused_gene_13 | |
| fused_gene_14 | |
| fused_gene_15 | |
| inversions_number | |
| inversions_gene_1 | |
| inversions_gene_2 | |
| inversions_gene_3 | |
| inversions_gene_4 | |
| inversions_gene_5 | |
| inversions_gene_6 | |
| inversions_gene_7 | |
| inversions_gene_8 | |
| inversions_gene_9 | |
| inversions_gene_10 | |
| translocations_number | |
| translocations_gene_1 | |
| translocations_gene_2 | |
| translocations_gene_3 | |
| translocations_gene_4 | |
| translocations_gene_5 | |
| translocations_gene_6 | |
| translocations_gene_7 | |
| translocations_gene_8 | |
| translocations_gene_9 | |
| translocations_gene_10 | |
| genomics_add_notes | Data imported via genomics package |
| gen_daf_note | |
| gen_dmf_note | |
| gen_lpf_note | |
| gen_flag_yn_1 | |
| gen_details_flag_1 | |
| gen_resolved_flag_1 | |
| gen_resp_flag_1 | |
| gen_flag_yn_2 | |
| gen_details_flag_2 | |
| gen_resolved_flag_2 | |
| gen_resp_flag_2 | |
| gen_flag_yn_3 | |
| gen_details_flag_3 | |
| gen_resolved_flag_3 | |
| gen_resp_flag_3 | |
| gen_unresolved_flag |
Table 2 - Output of genetex_to_redcap()
Import to REDCap®
The data in Table 2 is imported to REDCap® from the Shiny application by calling the function redcap_write_oneshot() from the REDCapR package. An example of a portion of that form with data imported from GENETEX is seen in Supplemental Figure 1.
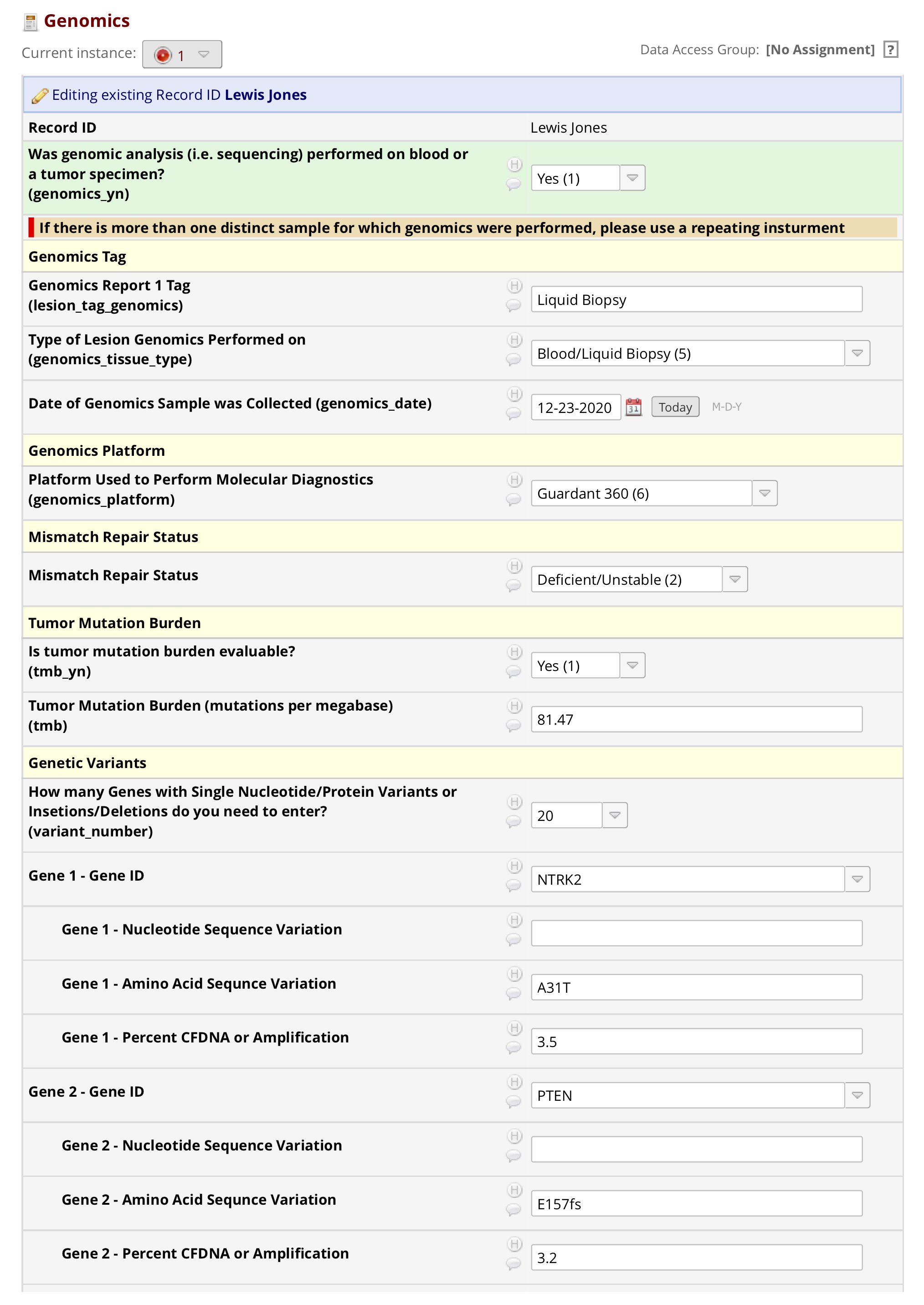
Supplemental Figure 1. Depcited is an example of the data form in REDCap® after the input clinico-genomic data has been passed to genetex_to_redcap().
Real-World Deployment
In order to evaluate the performance of augmented abstraction with GENETEX in the real-world setting compared to manual abstraction, we selected 7 genomic reports at random (3 Guardant, 2 Foundation Medicine, 1 MGH SNaPshot, 1 BWH Oncopanel) for abstraction. Each report was abstracted independently by two data abstractors via manual abstraction, as well as with GENETEX. In total, 744 data elements were captured from these 7 reports. Agreement rates between the two human abstractors was 99.19%. Importantly, >99% agreement was reached between both human abstractors and GENETEX (99.33% and 99.19%). Given that the agreement between classical and augmented abstraction was high, we next evaluated if the GENETEX pipeline would improve time-to-analysis. The mean time for manual abstraction for each report was 784.5 (range: 220.5-3096.5) seconds compared with 136 (range: 75- 216) seconds for augmented abstraction (Wilcoxon test p value = 0.015625).
Limitations and Solutions
GENETEX is to be used in conjunction with Genomics Instrument and thus, it is dependent on that form being installed into a REDCap® project. However, we have made the data dictionary freely available so that others may incorporate it into their individual project. Importantly, our regular expressions system of mapping variable names to key genomic data provides a high-degree of flexibility to map alternate variable names for REDCap® instruments with different data dictionaries. Additional limitations include the fact that at this time, GENETEX does not support all potential platforms available for CGD. However, due to its open-source position, external developers can perform pull-requests on GitHub for incorporation of additional platforms and future refinement. Lastly, genomic findings abstracted from individual reports describe results at the point in time at which they were provided are a historical record and may not represent the latest understanding of genomic science or precision oncology treatment paradigms.Conclusions
Ideally, structured data objects that exactly match clinical genomic reports would be universally available directly from the providers of genomic profiling reports, and may be at some point in the future. Until then, there may be a desire to abstract or otherwise process these reports into structured formats. GENETEX is a browser-based application for natural language processing of CGD obtained in routine clinical practice. It facilitates extraction of data from EHR, transformation of semi-structured data into a structured format and loading into REDCap®. Its Shiny extension enables non-R users to execute the package without familiarity of R. Real-world deployment of the GENETEX demonstrated excellent agreement with classical abstraction in roughly 1/5 of the time. Thus, augmented abstraction with browser-based applications can decrease the barrier to data capture and importantly improve time-to-analysis of clinico-genomic data.
License

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.